Variables in “Passive” Cryopreservative Methods: Standardizing Cell-Based Assays By Reducing Cryopreservation-Induced Variability
 Cells have become essential in modern medicine as therapies, vehicles for producing high‑value therapeutics, and tools for high‑throughput screening of pharmaceutical compounds. In the latter area, more than 50% of drug discovery screens use cell‑based assays, predominantly targeting receptors and ion channels using fluorescence‑based measurements, in either or both high‑ throughput and high‑content formats (1). Alongside a cell therapy market estimated to be worth some $5.0 billion by 2015 (2), the larger cell‑ based screening market is estimated to be worth $14.8.0 billion by 2018 (3).
Cells have become essential in modern medicine as therapies, vehicles for producing high‑value therapeutics, and tools for high‑throughput screening of pharmaceutical compounds. In the latter area, more than 50% of drug discovery screens use cell‑based assays, predominantly targeting receptors and ion channels using fluorescence‑based measurements, in either or both high‑ throughput and high‑content formats (1). Alongside a cell therapy market estimated to be worth some $5.0 billion by 2015 (2), the larger cell‑ based screening market is estimated to be worth $14.8.0 billion by 2018 (3).
Half of all screening groups express interest in using primary cells, stem cells, and early progenitor cells in their screens rather than cell lines (4). Such cells have close physiological similarities to cells found in actual tissues. That presents a range of challenges because such sources are known to be highly susceptible to batch‑to‑batch variability. A well‑ documented example is the use of human primary liver cells for hepatotoxicity studies (5). Hepatocytes are obtained from liver biopsies. They are dissociated and cryopreserved before being shipped to customers, where they are thawed and plated for cell‑based assays. Those cells do not proliferate, so new batches are required for extensive testing regimes. Because of inherent donor‑to‑donor variability (e.g., age, sex, lifestyle, and ethnicity), range of cell‑handling processes, differing cryopreservation techniques (6), and variations in reagents, results obtained from such screens can vary greatly. The limited access to suitable tissues also contributes to making such cells very limited and valuable.
Cryo- and Biobanking Processes
Stem and progenitor cells are actively being investigated for their ability to proliferate and consequently expand into large cell banks. That combined with their potential to be differentiated toward the lineage/ tissue of choice makes them ideal candidates for industrial‑scale screening operations (7). However, stem cells are prone to undergo phenotypic drift when expanded over long periods of time. Maintaining homogeneous stem cell banks requires the use of stringent methods through which all steps of the banking process (from establishing a master cell bank to generating the subsequent working cell banks) are controlled as much as possible (8). Wherever feasible, automation is recommended to remove human‑operator–induced variability (9). Homogeneous batches of cells are eventually stabilized over extended periods by cryopreservation techniques until such batches are used.
We and others have previously shown that the process by which cells are typically handled during manual cryopreservation operations significantly affected some fundamental properties of the cells upon thawing (e.g., proliferation). In this study, we wanted to investigate the effect of two cryopreservation approaches on the sensitization of the hepatic cell line HepG2 to methotrexate, as a simplified toxicity assay. We aimed to do that by comparing cells that had been frozen using either a standard laboratory “passive” freezing container in a mechanical freezer or a liquid‑nitrogen–based controlled‑rate freezer. We chose HepG2 because of their many phenotypic similarities with primary hepatocytes. They also minimize the inherent and unpredictable differences between batches of primary cells, which would otherwise affect the biological readouts of the assays.
Comparing Passive Freezing and Controlled-Rate Freezing
Plastic, alcohol‑filled containers are widely used and simple devices for freezing cells. The most common configuration consists of a foamy matrix surrounding a plastic holder holding two concentric rings of cryovials. The matrix is saturated with isopropanol at room temperature, the vials are loaded, and a closed container (Mr. Frosty from Nalgene) is placed in a mechanical freezer at –80°C overnight, generally for 24 hours. During this time, the cooling rate is assumed to be about –1 °C/min, at least in the early stages of the freezing process. The ease of using such a device and its cost efficiency makes it an attractive tool for low‑scale to mass cell cryopreservation.
At the other end of the cryopreservation technology spectrum are controlled‑rate freezers (CRFs). They consist of chambers and holding devices whose temperature is dynamically controlled through sensors and feedback loops, which allows for rapid and accurate temperature adjustments. These instruments are programmable, meaning that the actual freezing profile in the chamber can be customized for particular needs or cell types. That can be particularly useful if the optimal freezing rate turns out not to be –1 °C/min for a specific cell type.
Numerous reports have focused on the effects of cryopreservation methods on cells (10). The variables studied encompass the formulation of cryopreservative solutions, the effect of freezing rates on cell viability, subcellular and biochemical modifications, and so on. However, in those studies, what happens during the freezing process is treated as a given. The actual freezing profile (the variation of the freezing rate over time) that cells experience has not been precisely investigated. Reasons for this may be that temperature is principally measured in a cryochamber, not in the sample itself with the assumption that an alcohol container freezes all samples uniformly at a standard rate. The level of resolution needed for investigation is technically challenging: Cryovials are normally 1 mL in volume and do not easily accommodate temperature probes.
Freezing Cell Suspensions
We used a standard alcohol‑containing freezer (e.g., Mr. Frosty from Nalgene) accommodating 18 cryovials into two concentric rings of 12 and six holders respectively and a CRF (Kryo from Planer) to freeze cells. The alcohol‑filled container vials were placed in a –80 °C mechanical freezer. We placed vials to be frozen in the CRF in a metallic holder, itself placed in the CRF chamber, which was initially at room temperature as was the Mr. Frosty system. We optimized the chamber’s temperature profile to target a –1 °C/min sample freezing rate. Both devices had an identical number of cryovials.
We measured temperature profiles by inserting a thin thermocouple temperature probe into the cell suspension through a hole drilled in the cap of the cryovials and recorded the temperature every second. The cryopreservation medium consisted of 1 mL of culture medium containing 1 × 106 cells supplemented with 10% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO). Cell viability was >95% in all vials upon freezing.
Three types of samples were analyzed:
vials in the CRF frozen according to the programmed temperature profile
vials located in the outer ring of the alcohol‑filled container stored at –80 °C
vials located in the inner ring of the alcohol‑filled container stored at –80 °C.
We stored both batches of frozen cryovials at –80 °C for 24 hours (replicating a common scenario for cell‑culture laboratories, in which cryovials are frequently left overnight before being placed in liquid nitrogen). We repeated these experiments six times independently. For each of the three scenarios, we calculated the freezing rate of the samples over time from the temperature profiles. We further analyzed those freezing rates in terms of reproducibility by measuring the standard deviation of the six curves over time.
Cell Recovery and Toxicity Assessment
The effect of the freezing profile on cell function was tested using HepG2 cells, an immortal cell line derived from a human hepatocellular carcinoma. HepG2 are suitable models of hepatocytes and are often used to study some hepatic functions associated with liver cells (e.g., drug metabolism for in vitro toxicity studies) (11). In typical toxicity studies, cell proliferation and/or cell death are critical parameters to measure. This experiment replicates such a scenario and uses recovery and cell viability to assess the effect of the freezing profiles on our end point measurements.
Upon thawing the cells, we immediately placed them in the wells of a real‑time cell electrosensing plate (RT‑CES, Roche) and measured their proliferation over the first 24 hours. An RT‑CES plate consists of a 96‑well plate with the bottom of its wells covered with gold electrodes. Within those wells, an electrical current flows. Cells are deposited on the electrodes and incubated in medium at 37 °C in a 5% CO2 atmosphere. Live cells adhere to the electrodes, creating an impedance, which is measured and converted into an arbitrary cell index (CI) (12). This CI is linearly related to the cell number when the cells are in proliferation phase. So it increases with cell proliferation, eventually reaching a plateau when the bottom of a well is covered with a saturated monolayer of cells.
Plating efficiency (the percentage of live cells adhering to the culture flasks upon thawing) and subsequent cell growth are both affected by the quality of cell populations following a cryopreservation process (e.g., the number of live cells and their ability to adhere quickly to the substrate and divide rapidly when entering the proliferation regime). These combined cell properties (viability, adhesion, and growth upon thawing) are referred to as cell recovery in the following text. Impaired cells or populations of poorer quality display poorer recovery, which is reflected by CI curves over time.
We tested the response of the HepG2 cells to the exposure of hepatotoxic drug methotrexate (MTX) in a toxicology assay. The cells were exposed to a concentration of MTX chosen to kill approximately 50% of the cells within 24 hours, as preliminarily determined on healthy proliferating cells. The drug concentration required to kill 50% of healthy cells is also known as the EC50 of the drug, or effective concentration required to obtain 50% of an effect (cell death here). It is a benchmark concentration commonly used in pharmacology to assess the toxicity of compounds. At such concentration, fragile or damaged cells are more susceptible to toxic challenges and so have lower viability than their healthier counterparts. We used RT‑CES to monitor the viability of MTX‑treated cells.
Freezing Profiles
Figure 1a shows typical temperature profiles of the cell samples in each freezing system and the CRF chamber. The green curve shows the temperature of a cryovial frozen using the alcohol‑filled container at –80 °C in a mechanical freezer. The samples’ temperature variation is not subjected to a uniform freezing rate. Instead, it significantly differs from the expected –1 °C/min rate, gradually accelerating before the sample freezes, slowing down around the time when the sample freezes, accelerating again steeply after that point, and finally slowing down as the temperature reaches –80 °C.
The red curves show the same profile measured in a CRF optimized to target a freezing rate of –1 °C/min. The blue curve shows the measured‑temperature profile of the actual CRF chamber required to achieve the profile of the red sample curve. The slope of this profile is predominantly –1 °C/min in the first 45 min. The steep temperature drop followed by a mild positive gradient between 12 and 20 min were necessary to counteract the release of energy from the cryovials when the cell suspension suddenly freezes. It was also used to initiate the onset of freezing at a consistent point in the profile. Such release of energy happens when water suddenly turns to ice, and it is sufficient to trigger a much more noticeable temperature jump than the small bump observed in this example (red curve between 12 and 20 min).
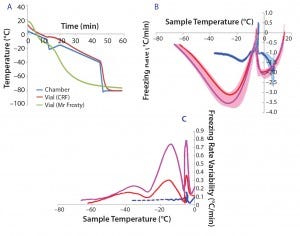
Figure 1: (a) Temperature profiles and sample freezing rates during cryopreservation in a Mr. Frosty or a controlled-rate freezer (CRF); (b) comparison of the freezing rates measured in both devices; blue represents the vials in the CRF, red and purple indicate vials in the inner and outer wells of the alcohol container respectively. The envelopes are standard deviations of the measurements, showing both the range and variability of the measurements.
After 45 min, the chamber temperature dropped to –80 °C because the cells were then definitely frozen, and the freezing rate was close to the nominal –1°C/min for long enough. Variability of the temperature probe was consistently 0.1 °C/min or less. Although the ideal freezing rate can be different for different cell types and must be optimized in each case, this example demonstrates the strong differences in temperature profiles experienced by cell samples between the two freezing devices used.
Because cell quality is known to be affected by freezing regime, it is important to implement reproducible cryopreservation processes to minimize variability of subsequent effects on cells. In six independent experiments, we further examined the freezing rates in the samples for vials stored either in the outer or inner wells of the alcohol‑filled container (Figure 1b, purple and red curves, respectively) and in a CRF (blue curve). We observed a positive spike in the freezing rate for all conditions around the freezing temperature (about –5 °C). Though fluctuating mildly, the CRF freezing rates remained much closer to the ideal –1 °C/min rates than those measured in the alcohol‑filled container. Figure 1c shows variability between independent runs of freezing rates achieved within sample vials. The greatest variability was measured for vials stored in the outer wells of a Mr. Frosty system, and then by those stored in the inner wells. From the freezing point to cooler temperatures, we observed three peaks in the alcohol container samples, indicating strong fluctuating freezing rates over time that were well above the 0.1 °C/min variability associated with the temperature probe. By contrast, freezing the cells using a CRF was a highly reproducible process, as exemplified with the minimal variability measured (blue curve in Figure 1c).
Table 1 summarizes the performance of both systems. Overall, the CRF was closer to the –1 °C/min target rate. The average freezing rate of the CRF was –1.07 °C/min before and after the freezing point. Only the vials in the outer wells of the alcohol container displayed on average a similar rate of –1.09 °C/min before the freezing point, though showing greater rates and variability (as shown by the standard deviations and coefficients of variation) after the freezing point and for vials placed in the inner wells of the device. The alcohol‑container freezing rates were approximately three times more variable than the CRF across the whole temperature range. Furthermore, the range of its freezing rate was about three times as large as the range for the CRF before reaching the freezing point and nearly six times as large after that point.
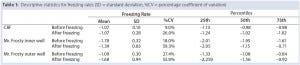
Table 1: Descriptive statistics for freezing rates (SD = standard deviation, %CV = percentage coefficient of variation)
Cell Viability and Function
We designed the cell recovery and toxicity assessment experiments to evaluate whether the significant differences in freezing patterns observed in the three types of samples would translate into measurable differences in cell performance upon thawing. We observed that the HepG2 cells with fastest recovery came from the CRF cryovials, followed by those stored in the inner wall of a Mr. Frosty container (Figure 2a). This recovery pattern showed a negative trend with the variability in freezing rates (Figure 1c). Such a trend was less marked in relation to the actual values of the freezing rates, although the overall higher rates experienced in the Mr. Frosty container resulted in poorer cell recovery for cells cryopreserved using this device. Those data suggest that the freezing rates before and after the freezing point, as well as the variability in freezing profiles experiences by the vials, all combine to affect cell recovery.
Finally, the hepatotoxicity test was carried out as a simulation of real life and industrially/clinically relevant drug screening application for such cells and storage conditions (Figure 2b). The viability from the CRF cells was indeed close to the targeted 50%, indicating that the cells were performing as if they were cultured fresh, not from a cryopreserved vial. However, the viability of the cells from inner and outer wells of the alcohol container device were much reduced (by 23% and 43%, respectively, compared with the viability measured on the CRF cells). This result indicated that these cells were significantly more fragile and therefore more sensitive to the MTX. The output of this assay also agrees with data from the proliferation assay (Figure 2a) and the variability in freezing rates (Figure 1c).
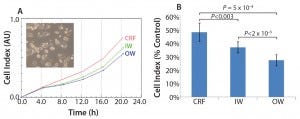
Figure 2: (a) HepG2 cell recovery and (b) susceptibility to toxic challenge following cryopreservation; CRF = controlled rate freezer, IW = inner wells and OW = outer wells of alcohol-filled container.
Comparisons
Given the stakes in the cell‑based industry, simple and passive cryopreservat ion techniques might not always provide an adequate enough level of control for optimal cell performances (13). Temperature profiles experienced by cells during cryopreservation can be highly variable from vial to vial as well as batch to batch. Such a high level of variability might have detrimental and significant consequences on cell behavior upon thawing.
Using a CRF can reduce that variability and could provide a near‑ optimal functionality profile for cells, which (along with an audit trail) is a desirable property in many industrial and clinical processes (14). Using a CRF also can facilitate the definition of cell product specifications and the implementation of tolerance levels for quality control and regulatory purposes. A programmable freezing profile also lends itself to further optimization of cryopreservation profiles, which could differ among different cell types and cryopreservative agents.
In the field of cell cryopreservation, a CRF currently provides the most robustly controlled platform needed to achieve accuracy, consistency, and reproducibility at critical stages of a cell‑banking process. However, a passive freezing device (e.g., alcohol filled container) has the advantages of economy and simplicity and can often be adequate for the job in hand.
References
1 Moeller TA, Shukla SJ, Xia M. Assessment of Compound Hepatotoxicity Using Human Plateable Cryopreserved Hepatocytes in a 1,536‑Well‑Plate Format. Assay Drug Dev. Technol. 10(1) 2012: 78–87.
2 Taking Stock of Regenerative Medicine in the United Kingdom: Research and Analysis. UK Department for Business, Innovation, and Skills, 18 July 2011, Ref : 11/1056; www.gov. uk/government/publications/regenerative‑ medicine‑in‑the‑uk‑taking‑stock.
3 Cell-Based Assays Market by Product (Reagents and Assay Kits (Cytotoxicity, GPCR), Cell Lines, Plate Readers, HCS, HTS, Software, and Assay Development Services), Application (Drug Discovery, ADMET), and End User (Pharmaceutical, CRO): Global Forecast to 2018. MarketsandMarkets: Chicago, IL; www. marketsandmarkets.com/Market‑Reports/cell‑ based‑assays‑market‑119917269.html.
4 Sartipy P, Björquist P. Concise Review: Human Pluripotent Stem Cell‑Based Models for Cardiac and Hepatic Toxicity Assessment. Stem Cells 29(5) 2011: 744–748.
5 McKim JM Jr. Building a Tiered Approach to In Vitro Predictive Toxicity Screening: A Focus On Assays with In Vivo Relevance. Comb. Chem. High-Throughput Screen. 13(2) 2010: 188–206.
6 Massie I, et al. Storage Temperatures for Cold‑Chain Delivery in Cell Therapy: A Study of Alginate‑Encapsulated Liver‑Cell Spheroids Stored at –80 °C or –170 °C for Up to 1 year. Tissue Eng. Part C Methods 19(3) 2013: 189–195.
7 Baxter MA, et al. Generating Hepatic Cell Lineages from Pluripotent Stem Cells for Drug Toxicity Screening. Stem Cell Res. 5(1) 2010: 4–22.
8 Thirumala S, Goebel WS, Woods EJ. Clinical‑Grade Adult Stem Cell Banking. Organogenesis 5(3) 2009: 143–154.
9 Williams DJ, et al. Precision Manufacturing for Clinical‑Quality Regenerative Medicines. Philos. Trans. A Math Phys. Eng. Sci. 370(1973) 2012: 3924–3949.
10 Perez‑Oteyza J, et al. Controlled‑Rate versus Uncontrolled‑Rate Cryopreservation of Peripheral Blood Progenitor Cells: A Prospective Multicenter Study. Haematologica 83(11) 1998: 1001��–1005.
11 Agren R, et al. Identification of Anticancer Drugs for Hepatocellular Carcinoma Through Personalized Genome‑Scale Metabolic Modeling. Mol. Syst. Biol. 10(3) 2014: 721.
12 Seiffert JM, et al. Dynamic Monitoring of Metal Oxide Nanoparticle Toxicity by Label‑Free Impedance Sensing. Chem. Res. Toxicol. 25(1) 2012: 140–152.
13 Li Y, Ma T. Bioprocessing of Cryopreservation for Large‑Scale Banking of Human Pluripotent Stem Cells. Biores. Open Access 1(5) 2012: 205–214.
14 Stéphenne X, Najimi M, Sokal EM. Hepatocyte Cryopreservation: Is It Time to Change the Strategy? World J. Gastroenterol. 16(1) 2010: 1–14.
Corresponding author Marc-Olivier Baradez is lead scientist analytical development at the Cell Therapy Catapult, 12th Floor Tower Wing, Guy’s Hospital, Great Maze Pond, London, SE1 9RT, UK; [email protected]. Tamara Lekishvili is a research scientist at LGC, Queens Road, Teddington, Middlesex, TW11 0LY, UK.
This work was sponsored by the UK National Measurement Office.
Marc-Olivier Baradez and Tamara Lekishvili
You May Also Like