- Sponsored Content
- Bioreactors
Cell Culture Media Fingerprinting: A Three-Tiered Approach

Cell culture media range from simple components to complex chemically defined mixtures. They may contain a number of chemical components, each with individual chemical properties — and any variation in which ingredients can affect cell culture processes and biological products being manufactured. Although simple substances can be identified easily using well-known classical methods such as Fourier-transform infrared spectroscopy (FTIR), analysis becomes more challenging when dealing with complex mixtures.
Section VII-3-C of the Q7A guidance from the International Council for Harmonisation of Technical Requirements of Pharmaceuticals for Human Use (ICH) requires at least one test to verify the identity of each batch of raw material in addition to the supplier’s certificate of analysis (CoA), provided that a biomanufacturer has a system in place to evaluate its suppliers (1). To deliver a high-quality product reproducibly, media manufacturers are expected to have mature quality control (QC) and manufacturing procedures of their own in place.
By pooling the knowledge, experience, and technical expertise of both biomanufacturers and their media-supply partners, the BioPhorum Raw Material Program workstream on media analytics has created a standardized fingerprinting test suite and three-tier approach to media fingerprinting (2). Our goals were to define the “best” tests to use, reduce unnecessary testing, align test standards across suppliers and biomanufacturers, and make case studies more accessible. It also should enable simplicity and standardization, facilitate appropriate and informative fingerprint testing for media and other noncompendial materials, improve qualification across suppliers, and provide for more straightforward and leaner investigations in the future.
Also as part of its Raw Material Program, BioPhorum developed a strategic framework for solving technical problems relating to managing raw materials in the biopharmaceutical industry (3). The approach to media fingerprinting described herein fits with that framework because it seeks to harmonize industry needs and analytical methods by providing guidance and recommendations. If our recommendations are followed, then duplicate testing (across media suppliers and biopharmaceutical manufacturers) could be reduced or even eliminated. That ultimately would help companies to optimize their laboratory resources, releasing resources for additional testing. Increasing knowledge about media will enable enhanced identification (ID) and understanding of the effects of variations in media components on biological products and processes.
This is an abridged and adapted version of the November 2022 BioPhorum white paper on this topic (2). For more context and detail, refer to the original publication online.
Problem Statement
Biopharmaceutical manufacturing processes have evolved to comply with good manufacturing practices (GMPs) and controls. Inadequate practices have been eliminated, such as by moving away from the use of chemically undefined raw materials such as sera, soy hydrolysates, and rice proteins in cell culture media. Media users cannot quantify every single medium component, however, and so must apply appropriate analytical techniques for documenting intended use and acceptance of controlled risk regarding some product variation. If the critical components of a medium are unknown, then a suboptimal source of raw material might be used that could lead to the loss of one or more product batches. Such events can be expensive and threaten both scheduling and market supply. Defined risks should be considered for products and processes, and then acceptable risk controls can be put in place to mitigate them.
Progress has been slow toward understanding the GMP readiness (e.g., data handling) and different levels of maturity among fingerprinting methods. Biomanufacturers need to perform at least one test to verify the identity of each batch of raw material in addition to using its supplier’s CoA (1). For these reasons, many end users simply duplicate the basic-ID fingerprinting tests already performed by media manufacturers as part of their internal QC processes. Such duplication does not add value to the medium fingerprint (much less to patients). The approach set out herein provides users with a way to identify alternative methods for fingerprinting and verifying the quality of cell culture media.
Key challenges we identified in developing this approach include a lack of standard guidance for fingerprinting cell culture media and associated raw materials. Current testing may be indicative of formulation or quality attributes but not process impact or fit for purpose. Current ID methods required to fingerprint media formulations are cumbersome and often require different analytical techniques: e.g., high-performance liquid chromatography (HPLC) and inductively coupled plasma mass spectrometry (ICP-MS).
Three-Tier Testing for Media Fingerprinting
The BioPhorum three-tier testing approach (Figure 1) guides users in choosing suitable methods for ID testing of incoming raw materials based on their chemical properties, the goals of media characterization, and available resources. This approach is intended to prevent duplicate testing and help ensure that media users can trust and use data provided by media manufacturers.
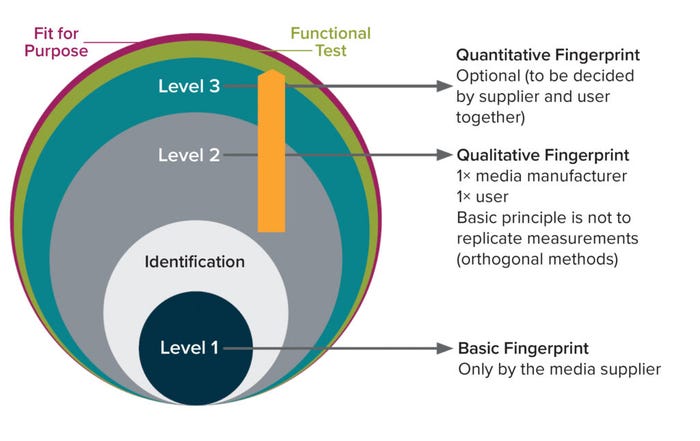
Figure 1: Three-tier testing approach to media fingerprinting (2).
In that context, it is the responsibility of media-supply partners to perform Level 1 basic (ID) testing and one type of Level 2 qualitative fingerprinting to confirm without discrepancies the identity/quality of media provided. End users are advised to perform orthogonal Level 2 qualitative fingerprinting to add dimensionality to the suppliers’ material characterization. Chosen methods should not be the same those used by the media-supply partners. Finally, based on process knowledge and risk assessment of the material, Level 3 quantitative fingerprinting for specific compounds might be warranted. That can be performed either by a media-supply partner as an additional service or by the end-user company, depending on resources and relationships (e.g., supplier quality agreements).
Below, we describe the expectations for each level in the three-tier testing approach. The overall aim is to confirm that cell culture media are fit for purpose and that the levels of fingerprinting applied are the result of process knowledge and requirements. The approach provides a recommendation for media fingerprinting while protecting the intellectual property of media-supply partners. Additional details and best practices for each test method described in each level can be found in the appendix of our original publication (2). When applying Level 2 qualitative and Level 3 quantitative fingerprinting, end users must align intellectual property aspects with those of the media-supply partners.
Level 1 Basic-ID Fingerprinting: Basic-ID fingerprinting identifies one medium from another at a gross level through simple measurements. What is described will not uniquely identify each chemically defined medium; rather, it enables differentiation of media types. Physicochemical test methods are used commonly for basic-ID fingerprinting. Such simple techniques provide confirm that media have been formulated accurately without gross errors in overall composition. Osmolality and pH are critical process parameters (CPPs) in cell culture that are defined and optimized to maximize productivity. Refractive index and conductivity are additional measurements that can be used for confirming the concentration and composition of media formulations. Methods for basic-ID fingerprinting are listed in the “Level One” sidebar, and best practices are detailed in the appendix of our original publication (2).
Level 2 Qualitative Fingerprinting: A number of analytical methods are generally qualitative for media fingerprinting and provide rapid results. Commonly chosen spectroscopic technologies including infrared (IR), Raman, two-dimensional (2D) fluorescence, and total reflection X-ray fluorescence (TXRF) spectroscopies can be combined with multivariate data analysis. The expense, complexity, and environmental impact of chromatographic techniques typically precludes their use for fingerprinting (4). However, some uses of LC-MS/MS for cell-culture media fingerprinting have been published (5, 6). As the technology evolves, we expect that wider use of chromatographic methods for Level 2 qualitative fingerprinting will be considered.
Spectroscopic methods give information about how analytes interact with electromagnetic radiation, enabling collection of specific “fingerprints” of chemical compounds or mixtures. IR is the most established such method in the pharmaceutical industry (7), followed by Raman (8), and both methods have demonstrated utility for identification of simple compounds. Li et al. applied Raman spectroscopy to fingerprinting of complex culture media for Chinese hamster ovary (CHO) cell culture (9). Kirdar et al. used near-infrared (NIR) spectroscopy for fingerprinting a number of raw materials (10). However, some limitations arise when such methods are applied to complex samples.
Also known as excitation–emission matrix (EEM), 2D fluorescence spectroscopy has been applied successfully to cell culture media fingerprinting, and its potential for use with complex raw materials also was demonstrated by Li et al. (11). The technology has been applied to cell culture media characterization, including quantification of different compounds, with correlation to process performance (12–15).
None of those spectroscopic methods can detect all compounds in a complex cell culture media sample, but they can provide good enough information to support a robust characterization or ID approach. Unfortunately, no clear library of references is yet available to provide the specific extent of analytes that can be covered by each method, but their advantages and drawbacks are now well understood (4, 16). Identifying the most appropriate method depends on your characterization goals, the chemical properties of materials sampled, and available expertise.
In theory, Raman and IR both can detect most chemicals. However, they detect only those compounds that are present at high concentration in a cell culture medium — which in most cases will be buffers and amino acids, predominantly. Low-level vitamins and trace elements are unlikely to be detected with either method. Both techniques are sensitive to interference from the presence of water; Raman is sensitive to intrinsic fluorescence, which may cause high baselines (16, 17). Simply put, Raman and/or IR fingerprinting will not perform as well with aqueous and high-fluorescence substances as is likely to be necessary.
Fluorescence-based fingerprinting takes advantage of fluorophores such as tyrosine, tryptophan, and vitamins B6 and B2 in cell culture media (15). Those fluorophores thus can act as “sensors” for detection of changes to chromophore concentration; to the presence of quenching agents, such as some trace metals (e.g., iron and copper); to pH and solvent polarity; and to sample viscosity. Thus, the fluorescence method is highly sensitive to changes in cell culture media, including among compounds at very low concentration, but it is not sensitive to all compounds.
Each spectroscopic method has unique properties, advantages, and challenges. Figure 2 should help with selection of a spectroscopic technique to use in fingerprinting cell culture media. IR or Raman (if available) can be used for simple or complex samples with low fluorophore content, especially to focus on the main compounds (generally glucose, some amino acids, and buffers). Raman measurements can be made through packaging, with handheld instruments available for measurement of powders as well as liquids (18–21). However, when used for powder characterization, Raman spectroscopy is sensitive to the presence of polymorphs (22, 23). IR also can also be used for powders but will be sensitive to their moisture content and thus is not recommended for hygroscopic samples — a property of many complex cell culture media powders (16).
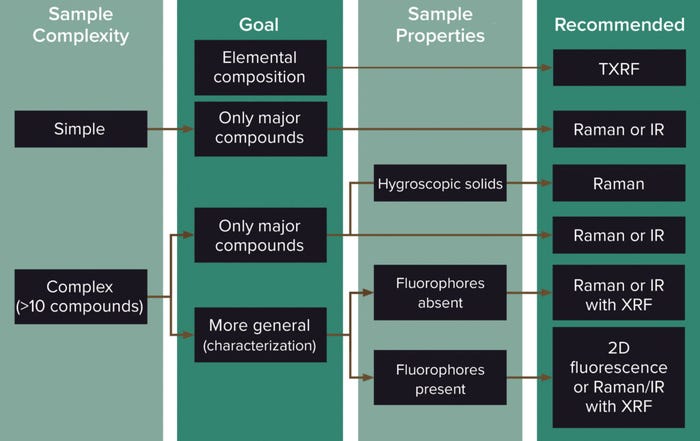
Figure 2: Decision-making chart for selecting a spectroscopic technique for fingerprinting cell culture media (2).
For highly complex samples, information about compounds in a larger range of chemical classes often will be sought. In such cases, 2D-fluorescence can be a good alternative because it detects changes in trace elements, vitamins, and amino acids at the concentrations typical in cell culture media or feeds (9, 11, 13–15, 24). However, complex cascade-like effects at play when variability is introduced in a medium make it difficult to predict fluorescence sensitivities. Adding or removing one component can start a chain reaction of fluorophores that can in turn affect the intensity of fluorescence.
Total reflection X-ray fluorescence spectroscopy (TXRF) can complement IR and Raman spectroscopy. It offers the ability to perform qualitative analysis of trace metals present in solubilized raw materials. The wavelength and energy level from the fluorescence radiation are specific to each trace element present in a sample. See below for more details. TXRF can be used either as a fast qualitative fingerprinting method for elemental profiling or as a quantitative fingerprint method.
Level 3 Quantitative Testing: When more extensive fingerprinting or investigative work is required, several methods can be applied, including TXRF, analytical chromatography, and ICP-MS. Below is a brief description of these techniques, with recommendations depending on the properties of a given cell culture medium. Other methods such as color spectroscopy and particle-size measurements are described in the appendix of our original publication (2).
Elemental Fingerprinting: ICP-MS is used to analyze individual elements in a sample according to their mass:charge ratio (25, 26). A sample usually is acidified and then introduced to the instrument, in which an aerosol is formed, and then ICP is used to form atomic ions that move into a mass spectrometer, where they are separated by mass and then moved to a detector. The elements are identified based on atomic mass, and a standard curve is used to quantify those of interest. The mass spectrum of a sample can be used as a fingerprint for comparison with other samples.
TXRF also can be used to perform quantitative analysis of trace elements detected in solubilized raw materials (liquid samples) (27). An internal standard is added to a sample, which is vortexed for mixing, and then a small amount of the sample is pipetted onto a glass disk and dried. Once analysis is complete, the concentration of each detected element is calculated either manually or automatically (using dedicated software connected to the instrument) based on the intensity of fluorescence radiation.
Like ICP-MS, TXRF can analyze solids through acid-digestion preparation of samples (28, 29). TXRF also provides rapid and direct qualitative and mass-ratio evaluation of elements present in only micrograms of liquid samples (without the chemical distortion of acid digestion), which indicates its utility for microanalytical applications. Another important advantage is that TXRF enables easy monitoring of bioprocesses through comparative study of their spectra and simultaneous qualitative inspection of atomic fingerprints from raw materials.
For liquid samples, the detection limits (DLs) of TXRF are competitive compared with other techniques: from hundreds of parts per trillion (ng/L) to tens of parts per billion (µg/L). ICP-MS is more sensitive to such samples, however, with DLs around 1 ppt (ng/L), and thus is more suitable for ultratrace analysis than any other technique. Note that it requires the performance of external calibration curves, whereas TXRF requires only the addition of an internal standard of known concentration (which is otherwise not present in the sample). Such a simplified quantification process brings important savings of time and cost.
Chromatography: Analytical chromatographic methods separate components of complex samples, with a number of detector technologies to identify and quantify analytes of interest: e.g., amino acids, vitamins, lipids, and other components that can be present at low concentration in cell culture media. HPLC and ultrahigh-performance liquid chromatography (UPLC) are used most often in reversed-phase mode with aqueous samples, but columns can use a breadth of chromatographic chemistries such as size exclusion, ion exchange (IEX), and affinity. Common detector options include those for ultraviolet/visible (UV/Vis), fluorescence, mass spectrometry (MS), refractive index (RI), conductivity, and charged aerosol (CAD). Analytes can be derivatized to enable or enhance detection. IEX can be used for analysis of buffers and other process liquids.
Users typically choose columns and detectors that are applicable to their analytes of interest; then develop a method based on parameters such as column size and format, mobile phase, column temperature, and sample volume; and run samples to generate chromatograms. The peaks in those images are measured and compared with a standard curve to quantify the analyte(s) of interest. Retention time of an unknown sample can be compared with the known reference standard to confirm its identity. For complex samples such as cell culture media, the chromatogram also provides a “fingerprint” that can be compared with other such data as another method of raw-material identification (5, 6).
If LC-MS is used, the mass spectrum adds another dimension to the fingerprint. A number of MS detectors are available. Figure 3 summarizes how methods from different detectors can be used for classical targeted/quantitative fingerprinting or for untargeted fingerprinting using multivariate data analysis. UV/Vis is a straightforward and inexpensive technique, but it can be subject to interferences in complex mixtures because it is not very specific. A good separation method is necessary. Derivatization methods, in which fluorescent or UV-absorbent labels are bound to analytes of interest, require more skill and equipment and are more time-consuming.
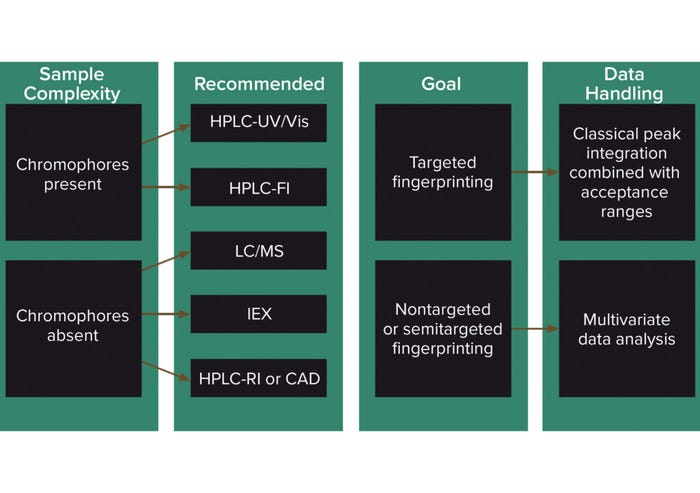
Figure 3: Decision chart for the selection of chromatographic techniques for the fingerprinting of cell culture media (2).
RI and CAD are ubiquitous methods and can detect organic compounds that UV or fluorescence detectors cannot (30). RI detectors are limited in sensitivity and cannot be combined with elution-gradient methods, which often are required for complex media samples. CAD detectors mostly are suited to detect nonvolatile compounds and are limited by narrow linear ranges. IEX can be used to screen charged compounds.
Using MS requires skilled method development but provides for quantitative screening of a wider range of analytes and gives a more informative fingerprint than other detection methods. LC-MS/MS is more selective and less prone to matrix effects than simple LC-MS for targeted fingerprinting (31). For nontargeted fingerprinting, no analytes of interest are specified, but the overall composition of a medium is monitored. In such cases, high-resolution LC-MS methods such as quadrupole time of flight (qToF) are recommended for reliable assignment of masses to the elemental composition of the media (32). That would allow compounds observed to be identified based on empirical evidence.
Figure 3 summarizes decision-making for chromatographic methods depending on analyte properties and whether the technique is intended to be used as a qualitative (nontargeted) or quantitative (targeted) tool.
An Analytical Toolbox
Although no “one-size-fits-all” approach to cell culture media fingerprinting is possible, we have outlined an approach in which “a few sizes fit most.” Application of the three-tier approach to characterization testing will benefit both media manufacturers and end-users. The type(s) of testing to use should be considered thoughtfully in the context of needs, goals, raw material properties, and risk.
It never has been easier to answer the question: “What type of fingerprinting testing should we do for our cell culture media?” Our original publication provides a context for decision-making to help standardize appropriate characterization testing (2). The analytical methods toolbox in its appendix lists fingerprinting tools common in the industry with comprehensive information to help users decide which tool best suits their needs. Included are advantages and disadvantages of each technique along with scientific principles and best-practice information. The toolbox also identifies media components that each technique measures.
By understanding and using the resources we have made available, both media manufacturers/suppliers and end users at biopharmaceutical companies can gain deeper insight into their cell culture media and product/process capabilities and requirements. That should help them focus their energies on what matters most in relation to raw materials: improved knowledge that can be applied to making lifesaving medicines better, safer, and more effective for patients.
References
1 ICH Q7. Good Manufacturing Practice Guidance for Active Pharmaceutical Ingredients. US Fed. Reg. 66(186) 2001: 49028–49029; https://database.ich.org/sites/default/files/Q7%20Guideline.pdf.
2 Talreja G, et al. Media Fingerprinting of Cell Culture Media: A Standardized Analytical Test Method Suite and Three-Tier Approach. BioPhorum: London, UK, November 2022; https://doi.org/10.46220/2022DS006.
3 Padmanabhan V. BioPhorum Raw Materials Strategic Framework. BioPhorum Operations Group: London, UK, July 2022; https://www.biophorum.com/download/raw-materials-strategy.
4 Rathore AS, Kumar D, Kateja N. Role of Raw Materials in Biopharmaceutical Manufacturing: Risk Analysis and Fingerprinting. Curr. Opin. Biotech. 53, 2018: 99–105; https://doi.org/10.1016/j.copbio.2017.12.022.
5 Krattenmacher F, et al. Effect of Manufacturing Temperature and Storage Duration on Stability of Chemically Defined Media Measured with LC-MS/MS. J. Chem. Technol. Biotechnol. 94, 2019: 1144–1155; https://doi.org/10.1002/jctb.5861.
6 Floris P, et al. Untargeted LC-MS/MS Profiling of Cell Culture Media Formulations for Evaluation of High Temperature Short Time Treatment Effects. Anal. Chem. 89, 2017: 9953–9960; https://doi.org/10.1021/acs.analchem.7b02290.
7 Morris KR, et al. Advances in Pharmaceutical Materials and Processing. Pharm. Sci. Technol. Today 1, 1998: 235–245; https://doi.org/10.1016/S1461-5347(98)00062-5.
8 Vankeirsbilck T, et al. Applications of Raman Spectroscopy in Pharmaceutical Analysis. Trends Anal. Chem. 21, 2002: 869–877; https://doi.org/10.1016/S0165-9936(02)01208-6.
9 Li, B. et al. Rapid Characterization and Quality Control of Complex Cell Culture Media Solutions Using Raman Spectroscopy and Chemometrics. Biotechnol. Bioeng. 107, 2010: 290–301; https://doi.org/10.1002/bit.22813.
10 Kirdar AO, et al. Application of Near-Infrared (NIR) Spectroscopy for Screening of Raw Materials Used in the Cell Culture Medium for the Production of a Recombinant Therapeutic Protein. Biotechnol. Progr. 26, 2010: 527–531; https://doi.org/10.1002/btpr.329.
11 Li B, et al. Fluorescence Excitation–Emission Matrix (EEM) Spectroscopy for Rapid Identification and Quality Evaluation of Cell Culture Media Components. Appl. Spectrosc. 65, 2011: 1240–1249; https://doi.org/10.1366/11-06383.
12 Groza RC, Calvet A, and Ryder AG. A Fluorescence Anisotropy Method for Measuring Protein Concentration in Complex Cell Culture Media. Anal. Chim. Acta 821, 2014: 54–61; https://doi.org/10.1016/j.aca.2014.03.007.
13 Ryan PW, et al. Prediction of Cell Culture Media Performance Using Fluorescence Spectroscopy. Anal. Chem. 82, 2010: 1311–1317; https://doi.org/10.1021/ac902337c.
14 Brunner M, et al. Towards Robust Cell Culture Processes: Unraveling the Impact of Media Preparation By Spectroscopic Online Monitoring. Eng. Life Sci. 19(10) 2019: 666–680; https://doi.org/10.1002/elsc.201900050.
15 Calvet A, Li B, Ryder AG. A Rapid Fluorescence Based Method for the Quantitative Analysis of Cell Culture Media Photo-Degradation. Anal. Chim. Acta 807, 2014: 111–119; https://doi.org/10.1016/j.aca.2013.11.028.
16 Ryder AG. Cell Culture Media Analysis Using Rapid Spectroscopic Methods. Curr. Opin. Chem. Eng. 22, 2018: 11–17; https://doi.org/10.1016/j.coche.2018.08.008.
17 Buckley K, Ryder AG. Applications of Raman Spectroscopy in Biopharmaceutical Manufacturing: A Short Review. Appl. Spectrosc. 71, 2017: 1085–1116; https://doi.org/10.1177/0003702817703270.
18 Arroyo-Cerezo A, et al. Deep (Offset) Non-Invasive Raman Spectroscopy for the Evaluation of Food and Beverages: A Review. LWT 149, 2021: 111822; https://doi.org/10.1016/j.lwt.2021.111822.
19 Coic L, et al. Evaluation of the Analytical Performances of Two Raman Handheld Spectrophotometers for Pharmaceutical Solid Dosage Form Quantitation. Talanta 214, 2020: 120888; https://doi.org/10.1016/j.talanta.2020.120888.
20 Mansouri MA, et al. Quantitation of Active Pharmaceutical Ingredient Through the Packaging Using Raman Handheld Spectrophotometers: A Comparison Study. Talanta 207, 2020: 120306; https://doi.org/10.1016/j.talanta.2019.120306.
21 Deidda R, et al. Vibrational Spectroscopy in Analysis of Pharmaceuticals: Critical Review of Innovative Portable and Handheld NIR and Raman Spectrophotometers. Trends Anal. Chem. 114, 2019: 251–259; http://dx.doi.org/10.1016/j.trac.2019.02.035.
22 Kent KP, Sahni M, Sharma C. Identification of Cell Culture Media with Raman Spectroscopy. SAFC: Lenexa, KA: https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/cell-culture-and-cell-culture-analysis/mammalian-cell-culture/cell-culture-media-with-raman-spectroscopy.
23 Strachan CJ, et al. Raman Spectroscopy for Quantitative Analysis of Pharmaceutical Solids. J. Pharm. Pharmacol. 59(2) 2007: 179–192; https://doi.org/10.1211/jpp.59.2.0005.
24 Li B, et al. Comprehensive, Quantitative Bioprocess Productivity Monitoring Using Fluorescence EEM Spectroscopy and Chemometrics. Analyst. 139, 2014: 1661–1671; https://doi.org/10.1039/C4AN00007B.
25 BioPhorum. Standards and Best Practices To Support Trace Metal Quantitation in Cell Culture Chemically Defined Media and Hydrolysates. US Pharmacopeial Convention: 2020; https://www.usp.org/events-training/course/standards-and-best-practices-support-trace-metal-quantification-cell-culture.
26 Mack A, Szorik A, Sharma C. DS Trace Elements Variation for Chemically Defined Cell Culture Media: Biopharmaceutical Industry Requirements and Cross-Company Collaboration To Mitigate Risks. BioPhorum: London, UK, March 2022; https://www.biophorum.com/download/trace-element-variation-for-chemically-defined-cell-culture-media-biopharmaceutical-industry-requirements-and-cross-company-collaboration-to-mitigate-risks.
27 Tibo Y, Xinyang F, Jinge Z. Total Reflection X-Ray Fluorescence Spectroscopy. Open Access Libr. J. 7(8) 2020: 1–12; https://doi.org/10.4236/oalib.1106671.
28 Fernández-Ruiz R. TXRF Spectrometry As a Powerful Tool for the Study of Metallic Traces in Biological Systems. Develop. Anal. Chem. July 2014: https://www.researchgate.net/publication/263951282_TXRF_Spectrometry_as_a_Powerful_Tool_for_the_Study_of_Metallic_Traces_in_Biological_Systems.
29 Mondia JP, et al. Using X-Ray Fluorescence To Measure Inorganics in Biopharmaceutical Raw Materials. Anal. Meth. UK 7(8) 2015: 3545–3550; https://doi.org/10.1039/C4AY02936D.
30 Bareford L, et al. Development of a Rapid and Reliable Analytical Method for Screening Poloxamer 188 for Use in Cell Culture Process. Biotechnol. Progr. 35(3) 2019: e2792; https://doi.org/10.1002/btpr.2792.
31 Sun Z, et al. High-Throughput LC-MS Quantitation of Cell Culture Metabolites. Biologicals 61, 2019: 44–51; https://doi.org/10.1016/j.biologicals.2019.07.003.
32 Martano G, et al. Fast Sampling Method for Mammalian Cell Metabolic Analyses Using Liquid Chromatography–Mass Spectrometry. Nat. Protoc. 10, 2015: 1–11; https://doi.org/10.1038/nprot.2014.198.
Gitanjali Talreja (Biogen); Amandine Calvet (Boehringer Ingelheim); Cheryl Fuchs (Merck & Co. Inc., Rahway, NJ, USA) are the authors of this work. Nadya Morales-Cumming (BASF); Mary Szorik (Cytiva); Divya Vasudevan (Roche/Genentech); Alaric Collins (GlaxoSmithKline); Jude Lakbub (Merck, Life Science Business Sector); Shawn Nelson (Procelys); Keith Freel (Thermo Fisher Scientific); Lesley Holt, Jannika Ilievska Kremer, Louisa Mitchell, and Jane Worthington (BioPhorum) all contributed. Contact Worthington at BioPhorum, The Gridiron Building, 1 Pancras Square, London N1C 4AG, United Kingdom; 44-7535-259-168; [email protected]; https://www.biophorum.com.
You May Also Like





