Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
Single-use concepts are widespread in all unit operations of the biopharmaceutical industry. Although single-use technology is rapidly advancing and considered to be highly advantageous in many regards (1,2,3), in some cases it cannot (yet) compete with classical manufacturing systems. Processes with a demanding character (e.g., high cell densities, high titers, high turbidities, increased particle/contaminant loads) especially can bring disposables to their limit of technical feasibility, especially in product harvesting (4,5,6).
Here we focus on that step, which is defined as a removal of cells, debris, and (ideally) typical contaminants such as host-cell proteins (HCPs) and DNA from process fluid. In conventional (multiuse) facilities, a typical harvest is a multistep procedure:
a centrifugation step for removal of cells and solids >1 µm
a depth filtration step for removal of smaller particles, colloids, and contaminants
a final filtration step to remove or reduce bioburden.
To perform similar tasks in a single-use facility, different approaches have been necessary because single-use centrifuges were not available until only recently. The most convenient way was to replace the centrifugation step by direct filtration of process fluid through a cascade of depth filters. In general, such an approach would partially remove contaminants and significantly reduce turbity beyond those filters. (7).
Although such filtration results may be promising, depth filtration does have several disadvantages. Depth-filtration system capabilities alone are often insufficient. Flow rates can drop dramatically due to an early pressure increase (5,8). Problems can occur: e.g., filter blocking and turbidity breakthrough. Depth filtration is the only remaining filtration technology that requires preflushing of filter material, which is costly and laborious. Finally, harvesting 1,000-L to 2,000-L volumes (currently the upper limit in single-use bioreactors) using this approach can be economically questionable because of relatively low filtration capacities (per m2 of filter).
All those disadvantages have led some people to search for different methods that can be operated in a single-use set-up without a centrifuge: sedimentation (5), tangential-flow filtration (9), flocculation (10,11), and body-feed filtration (12). And several attempts have been made to remove the clarification step and directly bind target molecules to an adsorptive material (13). One possibility is settling cells based on gravity alone — or with addition of substances that can enhance settling velocity by aggregation (e.g., Chitosan or DEAE). Although sedimentation does not require additional technical equipment, concerns arise regarding settling time and reproducibility of the process, which could adversely affect product quality. Substances added to enhance settling must be removed from the process fluid downstream, which adds complexity to a purification process (10,11).
Those alternative methods apply under specific process conditions and with certain products, but they are not universally applicable. Meanwhile, single-use centrifuges have become commercially available (2). Their application remains limited with respect to removal capacity, achievable g forces, and scalability.
We evaluated filtration performance of a new single-use module operated in body-feed mode using diatomaceous earth (DE) as a filter aid in combination with a dead-end filter. The filter aid is crucial. Over 10,000 different species of extinct and living diatom algae have been described (14). Their skeletons consist of an inert SiO2 structure that is unlikely to affect product properties. Of the US diatomite production of 0.82 million metric tons per year, 75% is produced for filtration applications (15).
The Celpure DE (Advanced Minerals Corporation) that we used is different from common DE used in the food and beverage industry. It is highly purified (96–99% SiO2 with very low contaminants) and certified according to USP-NF requirements (16). During Celpure production, impurities are removed before calcination and fusion onto a diatom surface (17). Consequently, the product can be used in biopharmaceutical production without regulatory restrictions.
We decided to test different types (e.g., particle size and permeability) and concentrations of DE as well as different cell lines and starting conditions (cell concentration, viability) to develop a robust method with general applicability. To improve filtration performance, we tested different pH reduction approaches. To enlarge particle size and prevent release of submicron particles, it has been proposed in literature to cause particle precipitation by a pH shift (to pH 4.3–5.5) before clarification (18,19). Moreover, pH reduction can lead to DNA/HCP precipitation in the final filtrate (20).
Despite the many different flocculants for cell harvest such as pDADMAC, Chitosan, DEAE described in the literature (21,22), we focused on low-pH precipitation alone. Finally, we compared DBF filtration with a pH-reduced DBF filtration with direct filtration through a depth filter. Filtration using other depth-filter modules with an equivalent process fluid have led to capacities of ~50 L/m2. Subsequently we also scaled up the reduced-pH DBF to 600 L to demonstrate practicability and robustness at larger scales. During all experiments, we monitored important parameters such as filtration capacity; trending pressures and flow rates; turbidity; lactate dehydrogenase (LDH) content; and IgG1, HCP, and DNA contents.
Preliminary Experiments: To determine key parameters such as the optimal DE type, concentration, or the pH value, we conducted preliminary experiments at laboratory scale. Summarizing them here introduces this new single-use filtration technique.
Each experiment tested 500-mL portions of different mammalian cell culture fluids (cell concentration, viability, CHO cell line). We supplemented those fluids with different types of Celpure DE from Lehmann and Voss and Company KG: C65 permeability (0.065 mDarcy), C100 permeability (0.1 mDarcy), C300 permeability (0.3 mDarcy), and C1000 permeability (1.0 mDarcy). The filter-aid vendor provides permeability information for the raw material (23). Based on solids that needs to be removed in a real process, such values can vary. By testing different variations (0.26, 0.51, 0.77, and 1.03), we determined an optimal concentration ratio of filter aid to wet biomass. The experiments involved crude harvest material (6.5 × 106 cells/mL, 40% viability, 7.8% wet biomass). The final preliminary experiment presented herein compared filtration performance at neutral and reduced pH conditions using a crude harvest material (13.6 × 106 cells/mL, 73% viability, 8.5% wet biomass).
All laboratory-scale experiments involved a down-scale filtration system with a filter area of 1.3 × 103/m2. Peristaltic pumps — Tandem 1081 type (Scilog Inc.) — charged the fluids to the different filtration devices. A Deltran single-use pressure sensor (Utah Medical Products) monitored feed pressure before each filtration. We used a TE4101 balance (Sartorius Stedim Biotech GmbH) to monitor flow rate and filtration volume. All experiments began with a constant flow rate of 300 L/m2/h. When the feed pressure reached 1.5 bar, we switched the filtration modus from a constant flow rate to constant pressure by reducing the feed rate. Filtration was terminated at 50% of remaining flow.
Diatomaceous Body-Feed Filter Module: Figure 1 depicts filter devices for the DBF. The capsule consists of two filter layers (nominal pore size 10 µm) and a total filtration area of 0.23 m2 in a polypropylene housing. One empty module weighs 5 kg. Up to 30 modules can be assembled in one device for scaling up. A universal stainless steel holder (also usable with different depth filters) compresses the filtration devices at 25 kN clamping pressure. For process-scale experiments, we used Celpure C300 filter aid.

Figure 1:
Depth-Filter Module: For comparison, we tested a single-use depth-filter module provided by Sartorius Stedim Biotech. It consisted of a double-layer material with an overall filter area of 0.98 m2 in a polypropylene housing similar to that of the DBF modules. It combined two DE-containing cellulose layers with 15 µm and 2 µm retention rates.
Primary Harvest Fluid: The culture fluid used in our process-scale experiments comes from a high–cell-density CHO fed-batch process involving a 1,000-L single-use bioreactor. The product is a well-characterized, commercially available IgG1 with an isoelectric point (pI) of pH 8.25. Because of our study’s complexity, we used the harvest in two portions on two different days (day 11 for the comparison study, day 12 for the scale-up study with the low-pH DBF). Day-11 crude harvest showed a viable cell concentration of 19.7 × 106 cells/mL and 97% viability. Day 12 viable cell concentration was 17.6 × 106 cells/mL and viability was 95%.
Process-Scale Experiments: For each experiment, we transferred a defined harvest volume (Table 1) to a single-use Flexel 3D LevMix mixing system (Sartorius Stedim Biotech FMT SAS) by a 520 U or 620 U peristaltic pump (Watson Marlow). Feed and clarified harvest vessel volumes were monitored with a floor balance (Mettler Toledo) and pressure was monitored online with a Press-N-050, BR pressure controller (PendoTECH). For our reduced pH experiments, we adjusted pH to 5.0 by adding 2 mmol/L acetic acid (Carl Roth GmbH and Company KG) to the crude harvest after filter-aid dosage. A Mettler Toledo pH meter with a conventional glass electrode monitored pH. Figure 2 shows a process flow schematic.
Table 1
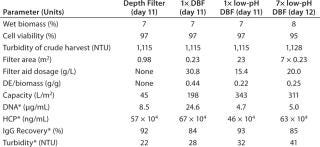
Table 1: 194;
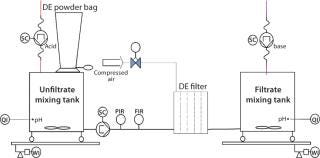
Figure 2:
We calculated the Celpure C300 dosage based on the determined wet biomass according to results of our preliminary experiments. After adding the filter aid through the mix-bag top port, we kept the suspension mixing at 140 rpm for two hours and then pumped it at a flow rate of 330 L/m2/h through the DBF filters into the filtrate receiving mix tank. For depth filtration, feed material was pumped out of the mix bag (without treatment) directly to the filtration membrane at ~60 L/m2/h.
After filtration terminated, we conducted a buffer wash using phosphate-buffered saline (PBS) with an osmolality of ~290 mosm/kg at a pH of 7.25. For the reduced-pH experiments, pH of the harvest fluid was adjusted to 7.0 with a 1 mol/L sodium hydroxide solution (Carl Roth). We took samples from the crude harvest, during filtration, and from different filtration pools: pool 1 (filtered harvest), pool 2 (after PBS flush), and pool 3 (after neutralization).
Turbidity Determination: We measured turbidity using a Hach 2100 laboratory turbidimeter that was calibrated with a Hach calibration standard before use each day. For this measurement we applied a sample volume of ~30 mL to sample cells before gently mixing and measuring.
DNA Determination: We determined DNA content using an Invitrogen Quant-iT PicoGreen assay from Life Technologies. Using this kit, 2-mL samples are centrifuged at 10,000g for five minutes directly after a sample is drawn (we used a Heraeus Multifuge X3R instrument from Thermo Fisher Scientific). Supernatant was stored at 2–8 °C until measurement for ≤14 days. Before measuring them, we allowed samples to equilibrate to room temperature, then measured them according to the assay instructions. The microplate was activated at 480 nm and read out at 520 nm by a Spectramax M5 instrument (Molecular Devices) using the SoftMax Pro software, version 5.01.
HCP Determination: We determined host-cell protein content using a F550 generic CHO enzyme-linked immunosorbent assay (ELISA) kit from Cygnus Technologies according to the supplier’s instructions. Samples were centrifuged for five minutes at 10,000g (again, with the Thermo Fisher Scientific Heraeus Multifuge X3R instrument), then stored at 2–8 °C for ≤14 days. Before measurement, we tempered the samples to room temperature for an hour, than incubated them with antibody and washed with Cygnus wash buffer. Extinction was measured at 450 nm by a Spectramax M5 instrument from Molecular Devices, again using SoftMax Pro software.
LDH and IgG1 Determination: Both LDH and IgG1 analysis involved a CuBiAn xc analyzer from Optocell Technology, now Roche Innovatis AG. We centrifuged 5-mL samples for five minutes at 10,000g (using the same instrument as above), then directly measured them. Depending on concentration, in some cases samples were diluted with PBS at pH 7.25.
LDH is an indicator for the break-up of mammalian cell cultures. We determined it using the LDH P-L assay (42.3– 1,150 U/L). IgG1 was determined using a high-sensitivity IgG1 assay (0.26–500.00 mg). We calibrated the analyzer before each use.
Wet Biomass Determination: We measured wet biomass in triplicate. A sample of 1-mL crude harvest was pipetted in a preweighed 2-mL Eppendorf cup and centrifuged for five minutes at 10,000g (with the same centrifuged identified above). After gently removing the supernatant we weighed the Eppendorf cup containing the cell pellet using an ML 204 analytical balance (Mettler Toledo).
Preliminary Experiments: Body-feed filtration is intended to remove cells effectively (even at high concentrations) and process-related impurities at the start of downstream processing. It can be an alternative to established single-use filtration methods that typically lack sufficient filtration capacity. Choosing the right raw material was crucial. DE composition, the geometric form of the diatoms, and the percentage of cracked diatomite are all important to filter-aid performance (24,25). Furthermore, characterization of DE in terms of permeability, wet density, surface area per gram, and relative retention rate is essential for predicting filtration characteristics.
We tested four grades of Celpure DE with different permeabilities. We determined filtration capacity and turbidity after the filtration process (Figure 3, TOP). Celpure C100 DE with 70–140 mDarcy provided the highest filtration capacity (374 L/m2), with turbidity found to be 55 NTU. Results showed that a too-low permeability (Celpure C65 DE, 40– 80 mDarcy) lowers filtration capacity (292 L/m2). Clarification results are good at 49 NTU.
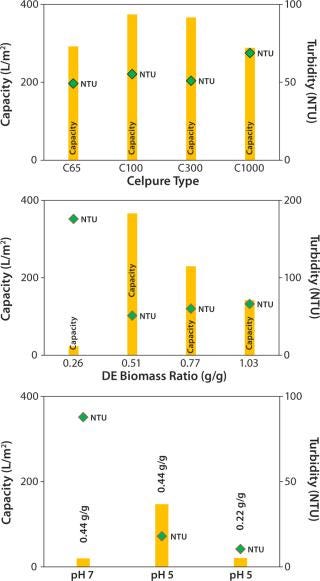
Figure 3:
The most suitable compromise came with results from Celpure C300 DE (150–300 mDarcy permeability) and a 366 L/m2 filtration capacity and clarification potential of 51 NTU. The experiment with the highest-porosity Celpure C1000 DE (750–1,250 mDarcy) gave the lowest filtration capacity (288 L/m2) and the highest turbidity (69 NTU). That filter aid builds a filtration cake with higher volume than the other Celpure types at the same concentration added to liquid. The filter unit is consequently filled with cake relatively quickly, which explains the lower filtration capacity. Although filtration throughput and quality depend on DE type, the amount of filter aid mixed with cell culture fluid also plays a major role. Figure 3 (CENTER) shows different ratios of the Celpure C300 filter aid to wet biomass (0.26– 1.03 g/g).
At a ratio of 0.26 g/g, the amount of filter aid is insufficient to allow build-up of a filter cake, but the required permeability provides a capacity of just 23 L/m2. At higher filter-aid concentrations (0.77 and 1.03 g/g), the filtration capacity becomes progressively limited, giving respective values of 230 and 142 L/m2. Turbidity in these cases was 60 and 66 NTU, respectively. Best results were obtained with a 0.51 g/g ratio of filter aid to wet biomass. We found the filtration capacity to be 366 L/m2 and turbidity to be the lowest of all tests (51 NTU).
To conclude, our evaluation of the optimal ratio between filter aid and wet biomass showed that too little filter aid does not enable effective build-up of a filter cake and that too much filter aid limits filtration capacity. Because this parameter significantly influences the cost of filtration, our aim was to minimize the amount of filter aid needed for attaining high performance. During supplementary laboratory-scale experiments (data not shown), fine-tuning the ratio to 0.44 at pH 7.0 improved results.
The approach of precipitation by pH reduction was considered to enhance clarification (26). Reduction of pH to 5.0 in cell culture broth induces impurity precipitation according to DLVO theory (27), which increases the average feed particle size (18,19). According to the theory, the charge of particulates in a strong ionic, polar solution (definitely the case with cell culture suspensions) prevents spontaneous agglomeration of particles and cells (27). By pH adjustment to acidic conditions, more positive surface charges evolve until the particles’ pI is reached. Thus, acidic conditions can improve agglomeration of particles and cells.
Compatibility of the reduced pH with the product’s pI must be proven. In our study, the MAb has a pI at pH 8.25, so the shift to acidic conditions can be performed without precipitation of the target protein. Figure 3 (BOTTOM) compares DBF under neutral pH conditions and reduced pH conditions. In these experiments, we stopped the filtration prematurely (as soon as pressure reached 1.5 bar) to allow for faster technology screening. So filtration capacities are lower. Nevertheless, results indicate that pH value affects filtration performance significantly. At pH 5.0, filtration capacity increases by a factor of 7 over that at pH 7.0; filtrate turbidity is reduced from 88 NTU at neutral pH to 18 NTU at reduced-pH conditions. That improvement allows for reduction of filter-aid dosage to only 50% of the starting value while maintaining a reasonably high filtration capacity and quality. According to these results, we operated the process-scale DBFs at pH 5.0 with a filter-aid dosage of only 0.22 g/g.
Process-Scale Experiments: We sought to verify our laboratory-scale results with process-scale testing to show the scalability of our method. Table 1 lists all process-scale experiments, including the most important starting values and results of the filtrations performed. On day 11 of the cell culture process, we used an aliquot of ~250 L to compare three filtration methods: depth filter, DBF, and low-pH DBF. On day 12 of the cultivation (with only slightly altered harvest conditions), the low-pH DBF was scaled up to ~650 L using seven process-scale modules.
Depth Filter: Pressure before the depth filter trends steadily higher until a sharp increase at a capacity of ~43 L/m2, indicating blockage of the filter membrane (Figure 4, TOP). At a final pressure of ~1 bar, filtration was terminated and a blow-out performed with 1-bar process air, followed by a 10-L PBS flush.
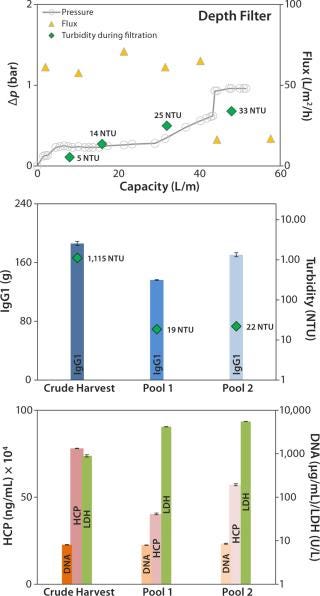
Figure 4:
The depth filter achieved lower flux rates (50 L/m2/h) than the DBF experiments. Filtration capacity was found to be 45 L/m2, and the achieved turbidity in the final harvest pool 2 (after the PBS flush) was 22 NTU (Figure 4, CENTER). But no significant reduction of contaminants released by the cells (DNA, HCP) was apparent (Figure 4, BOTTOM). Turbidity measured at different points from the clarified harvest stream showed an increase with forthgoing filtration capacity, but no filter breakthrough (Figure 4, TOP). Monitored LDH concentrations in the crude harvest and in the two pools (before and after the PBS flush) indicate moderate cell damage attributable to the applied filtration pressure. IgG1 recovery was high (92%).
Our depth-filter experiment confirmed results already achieved with other depth filters using the same process fluids (5). Using other process fluids with comparable cell concentrations, Pegel et al. reported filtration capacities of ≤150 L/m2 (8). That team’s results imply that this process fluid is a challenge to depth filters and that results can vary among processes even if key parameters (cell concentration) are similar.
Neutral-pH DBF: At neutral pH, the DBF allowed filtration of 198 L/m2 of crude harvest. The applied flux rate was high (~270 L/h/m2) until pressure reached a value >2 bar. The flow rate was manually reduced, and filtration was stopped when pressure repeatedly increased to 1.75 bar (Figure 5, TOP). Turbidity in the final pool 2 was measured at 28 NTU, and the turbidity trend during filtration (Figure 5, TOP and center) remained comparatively stable at ~30 NTU.
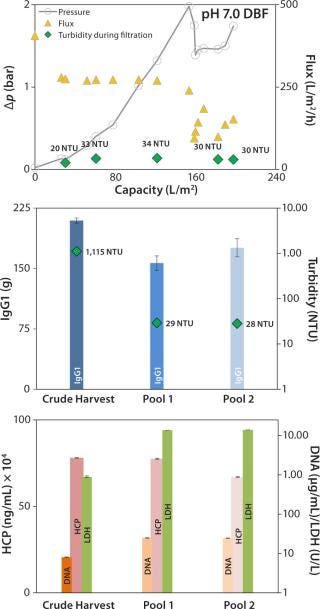
Figure 5:
No reduction of contaminants was obtained; only HCP values in the pool were reduced from 780 to 660 mg/mL. DNA and LDH values both increased by factors of 3 and 15.5, respectively (Figure 5, BOTTOM). That could come from cracked diatomite that is part of the filter-aid raw material even in low portions (25) and can disrupt cells during the filtration process. Alternatively, the filtration pressure in this experiment reached 2 bar, which could have intensified cell disruption. Recovery for this method was 84%, which is lower than the depth-filter value.
Reduced-pH DBF: In two experiments we used the DBF method combined with a reduced pH of 5.0:
for direct comparison with the other methods at day 11, using one process-scale DBF module
scale-up on day 12 of the cultivation with slightly altered crude harvest conditions (Table 1).
Figure 6 (TOP) depicts results of the onefold low-pH DBF, showing flux as well as the trends of pressure and turbidity in the clarified harvest stream. Pressure consistently increased during filtration. At 1.6 bar, we reduced the flow rate manually from ~330 L/m2/h to ~200 L/m2/h to prevent an overshoot. Then the pressure stabilized close to 1.6 bar, with no further increase detected. Turbidity during this filtration remained consistently low at 6–9 NTU (Figure 6, TOP). An overall capacity of 343 L/m2 was achieved, with final turbidity in pool 3 (after PBS wash and neutralization with sodium hydroxide) of 32 NTU (Figure 6, CENTER).
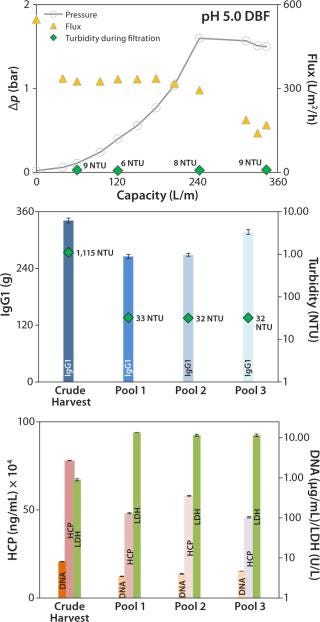
Figure 6:
For the scale-up experiment, we connected seven process-scale modules. Pressure trended lower here but was comparable to that in the DBF experiment above (Figure 7, TOP) without needing flux reduction to compensate for pressure. We terminated this filtration at 1.3 bar because of a lack of crude harvest. Flux remained very consistently at a high level (slightly above 300 L/m2/h). The overall capacity achieved was 311 L/m2, the final pool 3 exhibited a turbidity of 41 NTU (Figure 7, CENTER), and during filtration turbidity in the clarified harvest stream trended very low at 5–8 NTU (Figure 7, TOP).
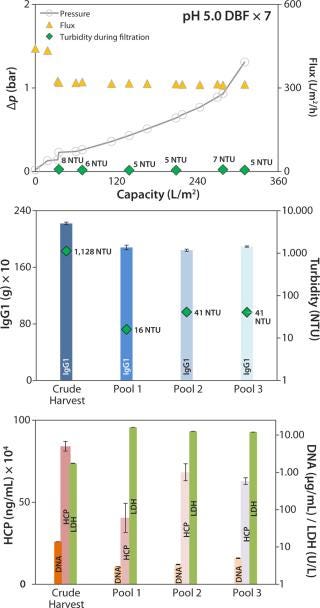
Figure 7:
IgG1 recovery was 85%, within an acceptable range (Figure 7, CENTER). HCP and DNA were reduced in both experiments: from 780 to 458 mg/mL and from 8.1 to 4.7 µg/mL (onefold low-pH DBF), and from 841 to 629 mg/mL and 13.8 to 5.0 µg/mL (sevenfold low-pH DBF), respectively. As in the 1× DBF experiment, LDH values were increased from crude harvest to pool 3 by a factor of 12.9 (onefold low-pH DBF) and a factor of 6.9 (sevenfold low-pH DBF), as Figure 7 (BOTTOM) shows. As discussed above for the depth-filter and neutral-pH DBF, filtration pressure is assumed to cause a certain degree of cell disruption. Beyond that, product recovery varied between 85% and 93%, which is in accordance with industry-standard depth filtrations (7,8).
We also assume that in the scale-up experiment with a low-pH DBF, the reduced postfiltration buffer flushing volume of only 11 L/m2 (compared with 15 L/m2 for the single low-pH experiment) caused a product loss that is a significant cost driver in overall purification. Adjusting pH to 7.0 in the final pool could be another reason for that product loss. Protein coils could have precipitated visibly for a temporary opaque appearance of the pool. Dosage of the caustic solution for neutralization to pH 7.0 probably may not have been as gentle as necessary. Thus, local pH extremes could have led to protein agglomeration. To improve this step, Sartorius Stedim Biotech is developing an integrated, ready-to-use process skid that will feature inline pH adjustment without overshooting.
Nevertheless, our tests showed excellent scalability for the DBF process compared with that obtained for depth-filter harvest methodologies, which had ≤50% differences reported by Yavorski et al. (28).
Preliminary Experiments: Body-feed filtration is intended to remove cells effectively (even at high concentrations) and process-related impurities at the start of downstream processing. It can be an alternative to established single-use filtration methods that typically lack sufficient filtration capacity. Choosing the right raw material was crucial. DE composition, the geometric form of the diatoms, and the percentage of cracked diatomite are all important to filter-aid performance (24,25). Furthermore, characterization of DE in terms of permeability, wet density, surface area per gram, and relative retention rate is essential for predicting filtration characteristics.
We tested four grades of Celpure DE with different permeabilities. We determined filtration capacity and turbidity after the filtration process (Figure 3, TOP). Celpure C100 DE with 70–140 mDarcy provided the highest filtration capacity (374 L/m2), with turbidity found to be 55 NTU. Results showed that a too-low permeability (Celpure C65 DE, 40– 80 mDarcy) lowers filtration capacity (292 L/m2). Clarification results are good at 49 NTU.

Figure 3:
The most suitable compromise came with results from Celpure C300 DE (150–300 mDarcy permeability) and a 366 L/m2 filtration capacity and clarification potential of 51 NTU. The experiment with the highest-porosity Celpure C1000 DE (750–1,250 mDarcy) gave the lowest filtration capacity (288 L/m2) and the highest turbidity (69 NTU). That filter aid builds a filtration cake with higher volume than the other Celpure types at the same concentration added to liquid. The filter unit is consequently filled with cake relatively quickly, which explains the lower filtration capacity. Although filtration throughput and quality depend on DE type, the amount of filter aid mixed with cell culture fluid also plays a major role. Figure 3 (CENTER) shows different ratios of the Celpure C300 filter aid to wet biomass (0.26– 1.03 g/g).
At a ratio of 0.26 g/g, the amount of filter aid is insufficient to allow build-up of a filter cake, but the required permeability provides a capacity of just 23 L/m2. At higher filter-aid concentrations (0.77 and 1.03 g/g), the filtration capacity becomes progressively limited, giving respective values of 230 and 142 L/m2. Turbidity in these cases was 60 and 66 NTU, respectively. Best results were obtained with a 0.51 g/g ratio of filter aid to wet biomass. We found the filtration capacity to be 366 L/m2 and turbidity to be the lowest of all tests (51 NTU).
To conclude, our evaluation of the optimal ratio between filter aid and wet biomass showed that too little filter aid does not enable effective build-up of a filter cake and that too much filter aid limits filtration capacity. Because this parameter significantly influences the cost of filtration, our aim was to minimize the amount of filter aid needed for attaining high performance. During supplementary laboratory-scale experiments (data not shown), fine-tuning the ratio to 0.44 at pH 7.0 improved results.
The approach of precipitation by pH reduction was considered to enhance clarification (26). Reduction of pH to 5.0 in cell culture broth induces impurity precipitation according to DLVO theory (27), which increases the average feed particle size (18,19). According to the theory, the charge of particulates in a strong ionic, polar solution (definitely the case with cell culture suspensions) prevents spontaneous agglomeration of particles and cells (27). By pH adjustment to acidic conditions, more positive surface charges evolve until the particles’ pI is reached. Thus, acidic conditions can improve agglomeration of particles and cells.
Compatibility of the reduced pH with the product’s pI must be proven. In our study, the MAb has a pI at pH 8.25, so the shift to acidic conditions can be performed without precipitation of the target protein. Figure 3 (BOTTOM) compares DBF under neutral pH conditions and reduced pH conditions. In these experiments, we stopped the filtration prematurely (as soon as pressure reached 1.5 bar) to allow for faster technology screening. So filtration capacities are lower. Nevertheless, results indicate that pH value affects filtration performance significantly. At pH 5.0, filtration capacity increases by a factor of 7 over that at pH 7.0; filtrate turbidity is reduced from 88 NTU at neutral pH to 18 NTU at reduced-pH conditions. That improvement allows for reduction of filter-aid dosage to only 50% of the starting value while maintaining a reasonably high filtration capacity and quality. According to these results, we operated the process-scale DBFs at pH 5.0 with a filter-aid dosage of only 0.22 g/g.
Process-Scale Experiments: We sought to verify our laboratory-scale results with process-scale testing to show the scalability of our method. Table 1 lists all process-scale experiments, including the most important starting values and results of the filtrations performed. On day 11 of the cell culture process, we used an aliquot of ~250 L to compare three filtration methods: depth filter, DBF, and low-pH DBF. On day 12 of the cultivation (with only slightly altered harvest conditions), the low-pH DBF was scaled up to ~650 L using seven process-scale modules.
Depth Filter: Pressure before the depth filter trends steadily higher until a sharp increase at a capacity of ~43 L/m2, indicating blockage of the filter membrane (Figure 4, TOP). At a final pressure of ~1 bar, filtration was terminated and a blow-out performed with 1-bar process air, followed by a 10-L PBS flush.

Figure 4:
The depth filter achieved lower flux rates (50 L/m2/h) than the DBF experiments. Filtration capacity was found to be 45 L/m2, and the achieved turbidity in the final harvest pool 2 (after the PBS flush) was 22 NTU (Figure 4, CENTER). But no significant reduction of contaminants released by the cells (DNA, HCP) was apparent (Figure 4, BOTTOM). Turbidity measured at different points from the clarified harvest stream showed an increase with forthgoing filtration capacity, but no filter breakthrough (Figure 4, TOP). Monitored LDH concentrations in the crude harvest and in the two pools (before and after the PBS flush) indicate moderate cell damage attributable to the applied filtration pressure. IgG1 recovery was high (92%).
Our depth-filter experiment confirmed results already achieved with other depth filters using the same process fluids (5). Using other process fluids with comparable cell concentrations, Pegel et al. reported filtration capacities of ≤150 L/m2 (8). That team’s results imply that this process fluid is a challenge to depth filters and that results can vary among processes even if key parameters (cell concentration) are similar.
Neutral-pH DBF: At neutral pH, the DBF allowed filtration of 198 L/m2 of crude harvest. The applied flux rate was high (~270 L/h/m2) until pressure reached a value >2 bar. The flow rate was manually reduced, and filtration was stopped when pressure repeatedly increased to 1.75 bar (Figure 5, TOP). Turbidity in the final pool 2 was measured at 28 NTU, and the turbidity trend during filtration (Figure 5, TOP and center) remained comparatively stable at ~30 NTU.

Figure 5:
No reduction of contaminants was obtained; only HCP values in the pool were reduced from 780 to 660 mg/mL. DNA and LDH values both increased by factors of 3 and 15.5, respectively (Figure 5, BOTTOM). That could come from cracked diatomite that is part of the filter-aid raw material even in low portions (25) and can disrupt cells during the filtration process. Alternatively, the filtration pressure in this experiment reached 2 bar, which could have intensified cell disruption. Recovery for this method was 84%, which is lower than the depth-filter value.
Reduced-pH DBF: In two experiments we used the DBF method combined with a reduced pH of 5.0:
for direct comparison with the other methods at day 11, using one process-scale DBF module
scale-up on day 12 of the cultivation with slightly altered crude harvest conditions (Table 1).
Figure 6 (TOP) depicts results of the onefold low-pH DBF, showing flux as well as the trends of pressure and turbidity in the clarified harvest stream. Pressure consistently increased during filtration. At 1.6 bar, we reduced the flow rate manually from ~330 L/m2/h to ~200 L/m2/h to prevent an overshoot. Then the pressure stabilized close to 1.6 bar, with no further increase detected. Turbidity during this filtration remained consistently low at 6–9 NTU (Figure 6, TOP). An overall capacity of 343 L/m2 was achieved, with final turbidity in pool 3 (after PBS wash and neutralization with sodium hydroxide) of 32 NTU (Figure 6, CENTER).

Figure 6:
For the scale-up experiment, we connected seven process-scale modules. Pressure trended lower here but was comparable to that in the DBF experiment above (Figure 7, TOP) without needing flux reduction to compensate for pressure. We terminated this filtration at 1.3 bar because of a lack of crude harvest. Flux remained very consistently at a high level (slightly above 300 L/m2/h). The overall capacity achieved was 311 L/m2, the final pool 3 exhibited a turbidity of 41 NTU (Figure 7, CENTER), and during filtration turbidity in the clarified harvest stream trended very low at 5–8 NTU (Figure 7, TOP).

Figure 7:
IgG1 recovery was 85%, within an acceptable range (Figure 7, CENTER). HCP and DNA were reduced in both experiments: from 780 to 458 mg/mL and from 8.1 to 4.7 µg/mL (onefold low-pH DBF), and from 841 to 629 mg/mL and 13.8 to 5.0 µg/mL (sevenfold low-pH DBF), respectively. As in the 1× DBF experiment, LDH values were increased from crude harvest to pool 3 by a factor of 12.9 (onefold low-pH DBF) and a factor of 6.9 (sevenfold low-pH DBF), as Figure 7 (BOTTOM) shows. As discussed above for the depth-filter and neutral-pH DBF, filtration pressure is assumed to cause a certain degree of cell disruption. Beyond that, product recovery varied between 85% and 93%, which is in accordance with industry-standard depth filtrations (7,8).
We also assume that in the scale-up experiment with a low-pH DBF, the reduced postfiltration buffer flushing volume of only 11 L/m2 (compared with 15 L/m2 for the single low-pH experiment) caused a product loss that is a significant cost driver in overall purification. Adjusting pH to 7.0 in the final pool could be another reason for that product loss. Protein coils could have precipitated visibly for a temporary opaque appearance of the pool. Dosage of the caustic solution for neutralization to pH 7.0 probably may not have been as gentle as necessary. Thus, local pH extremes could have led to protein agglomeration. To improve this step, Sartorius Stedim Biotech is developing an integrated, ready-to-use process skid that will feature inline pH adjustment without overshooting.
Nevertheless, our tests showed excellent scalability for the DBF process compared with that obtained for depth-filter harvest methodologies, which had ≤50% differences reported by Yavorski et al. (28).
The aim of our study was to demonstrate the universal applicability of a new single-use harvest method for mammalian cell culture streams, even at worst-case conditions. Testing with different cell lines and culture conditions allowed us to determine an optimal filter-aid concentration in relation to wet biomass, which is an easily accessible value for process-specific harvest processing. With respect to process economics, 50% reduction of filter aid needed for low-pH filtrations is promising.
Generally, the results of our process-scale experiments reinforced the results from DBF filtrations at laboratory scale, with reduced-pH experiments providing the best performance in terms of filtration capacity, flux, and contaminant removal. The applicable flux rate of this technology is advantageous: A harvest of 600 L was processed within only an hour of filtration using only seven modules. With 33 modules occupying the full space available space in the module holder, a theoretical harvest volume of ~3,000 L could be filtered in the same time.
This method allows filtration of high–cell-density processes in a single-use set-up without centrifugation for volumes found in single-use applications (≤2,000 L). Even challenging harvest solutions were filterable, with high flow rates and potentially significant reduction of contaminants. Following a linear approach, we found that scalability — one of the most important issues in bioprocessing — was easily attainable with very consistent process performance.
Author Details
Corresponding author Benjamin Minow, Dipl. Ing. (FH), is director of cell culture disposable manufacturing, and Florian Egner, Dipl. Ing. (FH), is a process engineer at Rentschler Biotechnologie GmbH, Erwin Rentschler Straße 21 88471 Laupheim, Germany, [email protected]. Franziska Jonas, Dipl. Ing. (FH), and Bertille Lagrange are with Sartorius Stedim Biotech GmbH, August Spindler Straße 11, 37079 Göttingen, Germany, [email protected].
REFERENCES
1.) Eibl, R, and D Eibl. 2009. Application of Disposable-Bag Bioreactors in Tissue Engineering and for the Production of Therapeutic Agents. Adv. Biochem. Eng. Biotechnol.:183-207.
2.) Shukla, AA, and U Gottschalk. 2013. Single-Use Disposable Technologies for Biopharmaceutical Manufacturing. Trends Biotechnol. 31:147-154.
3.) Zheng, R 2010. The Game Changer. BioProcess Int. 8:S4-S9.
4.) Lee, B, E Langer, and R Zheng Eibl, R and D. 2011.Next Generation Single-Use Bioreactor Technology and the Future of Biomanufacturing: A Summary from the Manufacturers and Users PerspectiveSingle-Use Technology in Biopharmaceutical Manufacture, Wiley-VCH Verlag GmbH, Weinheim:183-194.
5.) Minow, B, P Rogge, and K Thompson. 2012. Implementing a Fully Disposable MAb Manufacturing Facility. BioProcess Int. 10:48-57.
6.) Wagner, R, and D Mueller Subramanian, G. 2012.Full Plastics: Consequent Evolution in Pharmaceutical Biomanufacturing from Vial to WarehouseBiopharmaceutical Production Technology, Wiley-VCH:745-767.
7.) Yigzaw, Y. 2006. Exploitation of the Adsorptive Properties of Depth Filters for Host Cell Protein Removal During Monoclonal Antibody Purification. Biotechnol. Progr. 22:288-296.
8.) Pegel, A. 2011. Evaluating Disposable Depth Filtration Platforms for MAb Harvest Clarification. BioProcess Int. 9:52-56.
9.) Reis van, R. 1991. Industrial Scale Harvest of Proteins from Mammalian Cell Culture By Tangential Flow Filtration. Biotechnol Bioeng. 38:413-422.
10.) Schirmer, EB. 2010. Primary Clarification of Very High-Density Cell Culture Harvests By Enhanced Cell Settling. BioProcess Int. 8:32-39.
11.) Riske, F. 2007. The Use of Chitosan As a Flocculant in Mammalian Cell Culture Dramatically Improves Clarification Throughput Without Adversely Impacting Monoclonal Antibody Recovery. J. Biotechnol. 128:813-823.
12.) Delavaille, D. 2010.. How to Apply Old Techniques to New Processes.
13.) Anton, F. 2007.. Application of Direct Capturing in Downstream Processing of Proteins Using Membrane Based Chromatography (poster).
14.) Antonides, LE 1997. Diatomite. US Geological Survey. Mineral Commodity Summaries 24:1-4.
15.) Crangle, RD 2013. Diatomite: Statistics and Informationl. US Geological Survey. Mineral Commodity Summaries http://minerals.usgs.gov/minerals/pubs/commodity/diatomite:52-53.
16.) Purified Siliceous Earth 1948.USP35–NF30 2012.
17.) AMC10 2007.Advanced Minerals Regulatory Support Package: Celpure C, Version 1.8, Sartorius Stedim Biotech, Goettingen.
18.) Brodsky, Y. 2012. Caprylic Acid Precipitation Method for Impurity Reduction: An Alternative to Conventional Chromatography for Monoclonal Antibody Purification. Biotechnol. Bioeng. 109:2589-2598.
19.) Westoby, M. 2011. Effects of Solution Environment on Mammalian Cell Fermentation Broth Properties: Enhanced Impurity Removal and Clarification Performance. Biotechnol. Bioeng. 108:50-58.
20.) Yigzaw, Y 2008.. Improving Upstream Feed Stock to Downstream Operations.
21.) Salehizadeh, H, and SA Shojaosadati. 2001. Extracellular Biopolymeric Flocculants: Recent Trends and Biotechnological Importance. Biotechnol. Adv. 19:371-385.
22.) Roush, DJ, and Y Lu. 2008. Advances in Primary Recovery: Centrifugation and Membrane Technology. Biotechnol. Progr. 24:488-495.
23.) Technical Note AMC02, version 3.5. Comparing Conventional Diatomite and Celpure® Filter Aids.
24.) Begerow Product Line: User Information (in German), Eaton Technologies GmbH, Altlußheim.
25.) Hebmüller, F. Einflussfaktoren auf die Kieselgurfiltration von Bier.
26.) Thömmes, J, and M Etzel. 2007. Alternatives to Chromatography Separations. Biotechnol. Progr. 23:42-45.
27.) Gösele, W 2004.Handbuch der Mechanischen Fest-Flüssig-Trennung, Vulkan Verlag GmbH, Essen:178-180.
28.) Yavorsky, D. 2003. The Clarification of Bioreactor Cell Cultures for Biopharmaceuticals. Pharmaceut. Technol.:62-76.
You May Also Like