Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
Sponsored by GE HealthCare Technologies
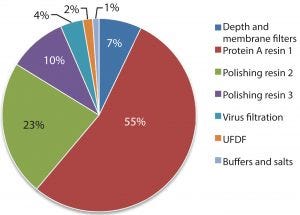
Figure 1: Estimated percentage of raw material costs for monoclonal antibody downstream process
Next-generation, high-capacity, alkali-stable protein A resins have recently become available in anticipation of increased production and throughput demand for protein therapeutic processes. Efficient caustic cleaning and bioburden control regimens have allowed agarose-based, alkali-stable protein A chromatography resins to become the backbone of many commercial purification processes for over a decade and have served as the primary capture step for monoclonal antibody (MAb) purification processes. However, the relatively high raw material costs, along with limitations to binding capacity and aggressive resin stripping strategies with current resins, still represent a high contribution to overall manufacturing cost of goods. To address this, a novel higher capacity prototype protein A resin from GE Healthcare (MabSelect™ PrismA) was evaluated using a CHO-expressed MAb. Stability and performance of the resin over its usable lifetime was studied by monitoring dynamic binding capacity (DBC), protein A ligand leachate, and total protein carryover.
Protein A is a 42 kDa protein found on the cell wall of Staphylococcus aureus. Staphylococcal protein A (SpA) is composed of five IgG-binding domains (E, D, A, B, and C) folded into a triple helix bundle. All domains have affinity for, and can specifically bind to the Fc region of antibodies. It is also known that multiple domains have affinity for the antigen-binding region (Fab) of the antibody, most notably the D domain (VH3 family). Many companies have purposely modified and mutated these domains to ensure high stability, specificity, and consistency of affinity purifications over the lifetime of the resin. These modified domains, due to their increased stability, have been produced as repeating subunits (repeating tetramer, pentamer, or hexamer) to further increase efficiency in binding capacity.
Protein A resin accounts for a large portion of the overall raw material costs to MAb processes, (Figure 1) and to a significant portion of overall operating costs (1) and cost of goods manufactured (COGM) (2). To drive down total COGM, either the raw material price must be lower or higher raw material utilization must be employed.
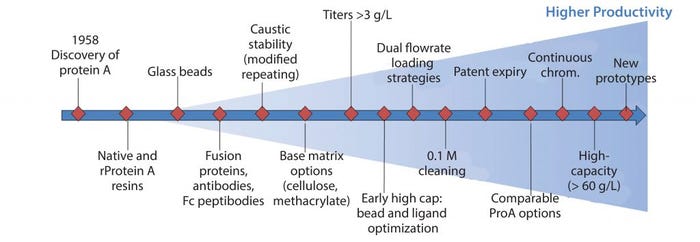
Figure 2: Protein A step schematic of improvements to process, equipment, and materials to meet demands of higher productivity
Advancements in fermentation and cell culture technologies have resulted in significant upstream productivity gains, thereby creating potential bottlenecks to downstream processing. To mitigate these potential bottlenecks, many technological advances in cell harvesting and capture strategies have been developed, optimized, and implemented. Capacity bottlenecks at the protein A step have been addressed through multifaceted improvements in process conditions (e.g., dual flowrate loading), equipment strategies (e.g., continuous chromatography), and most important, the protein A ligand and bead technologies (base matrix, ligand, bead size) (Figure 2). As a result, novel protein A resins are now commercially available and claim to have increased both alkaline stability for better microbial control and capacity (>60 g MAb/L resin).
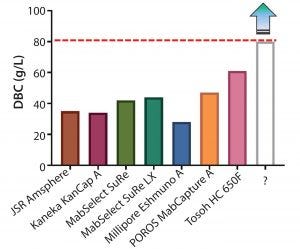
Figure 3: Dynamic binding capacities (3-minute residence time) of various commercially available protein A resins
Although multiple novel alkali-stable protein A resins are now available, dynamic binding capacities can vary widely (Figure 3), and the bar to update established platforms or modify existing commercial processes may be high. To justify a change from existing protein A resins, the following baseline criteria were considered: >50% increase in DBC, caustic stability for over 150 cycles with minimal drop in DBC, high product recovery (>90%), low protein A leachate profiles, and low protein carryover (lot-to-lot). GE Healthcare has produced a new prototype (MabSelect PrismA) that has a smaller particle size (d50) of about 60 μm, further alkaline stabilized protein A-derived ligand, and optimized bead characteristics (porosity).
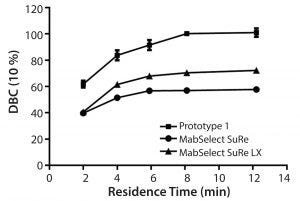
Figure 4: Dynamic binding capacity (10% breakthrough) as a function of residence time for MabSelect SuRe, MabSelect SuRe LX, and prototype (MabSelect PrismA)
Protein A Evaluation Using MabSelect SuRe, MabSelect SuRe LX, and Prototype (MabSelect PrismA) Dynamic Binding Capacity Assessment:
Comparison DBC studies were performed on MabSelect SuRe™, MabSelect SuRe LX, and the prototype (MabSelect PrismA) at various residence times (Figure 4). At 8 minute residence time, the observed DBCs plateau at about 50, 60, and 100 g/L for MabSelect SuRe, MabSelect SuRe LX, and the prototype (MabSelect PrismA), respectively. The prototype (MabSelect PrismA) DBC also was 50% higher in DBC at the lowest residence time of 2 minutes, whereas MabSelect SuRe and MabSelect SuRe LX appear to converge to the same value.
Cleaning and Sanitization Efficiency: Because of the elevated capacities observed for the prototype (MabSelect PrismA), cleaning and sanitization procedures were assessed for the potential increase in protein carryover. To assess cleaning and carryover, aged resin was generated by consecutive MAb protein load cycles at 80% DBC (4 minute residence time). A control mock elution was performed without cleaning in place (CIP) to assess total potential carryover before caustic treatment. Resin comparisons were assessed by cleaning first with mild caustic solution (25 mM sodium hydroxide), a mock elution (elution buffer only), multiple sequentially increasing caustic cleaning solutions, and finally a denaturing/ reducing solution of DTT/urea to assess further requirements for sanitization procedures.
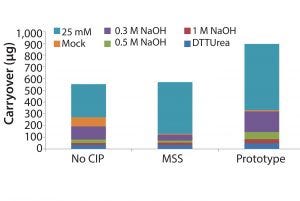
Figure 5: Carryover in total protein as measured by quantitative fluorescence of cleaning and sanitization fractions: 25 mM NaOH, mock elution buffer, 0.3 M NaOH, 0.5 M NaOH, 1 M NaOH, and DTT/urea solution.
Results indicate that significant residual protein was left bound to the resin with no CIP. Mild caustic cleaning (25 mM NaOH strip) is also insufficient to fully regenerate the resin. Prototype (MabSelect PrismA) resin was observed to have significantly more residual material as measured by total quantitative fluorescence and is most likely due to the increased loading of harvested cell culture fluid (Figure 5). Optimized cleaning and sanitization procedures may be required at these elevated loading, however, it appears that <100 μg of material was left over during 1.0 M NaOH and DTT/urea fractions combined.
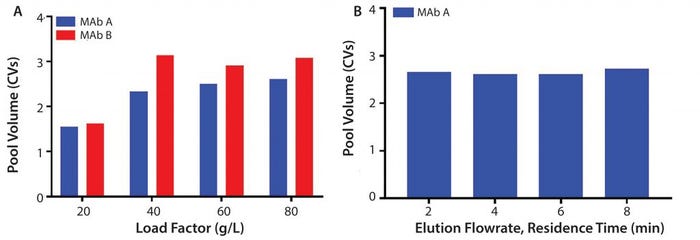
Figure 6: (A) Normalized pool volume as a function of load factor (g/L) for MAb A and Mab B; (B) elution flowrate (residence time, min) at 80 g/L MAb A
Process Performance and Resin Lifetime Studies: Several studies were executed to assess process performance (pool volume) on the prototype (MabSelect PrismA) and resin lifetime stability between resins. Pool volumes were evaluated for multiple MAb molecules on the prototype at protein loadings of 20, 40, 60, and 80 g/L. Results show DBC plateaus around 60 g/L with pool volumes of around 2.5–3 CV (column volume) (Figure 6a). Pool volumes also were evaluated at 80 g/L and varied elution residence times of 2, 4, 6, and 8 minutes. Experiments show consistent pool volumes of around 2.5 CV at the maximum capacity of 80 g/L (Figure 6b).
To assess the stability of protein A resin over its lifetime, cycling studies were performed comparing both MabSelect SuRe and prototype (MabSelect PrismA) resins using 0.3 M and 1.0 M NaOH sanitization solutions. Dynamic binding capacity was monitored both with and without protein load over 150 cycles to assess the effect of caustic stability and protein load independently. To eliminate the potential contribution to degradation from protein loading, cycles without protein load were performed by skipping the load step and directly washing the column after equilibration.
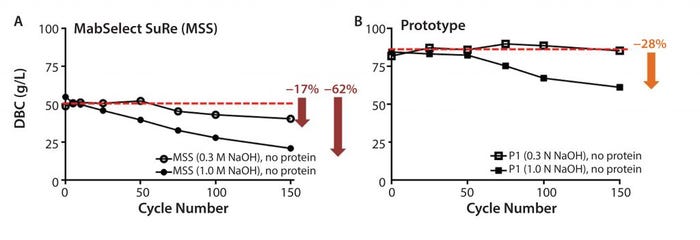
Figure 7: Resin lifetime study comparison between (a) MabSelect SuRe resin (no protein load step) and (b) prototype (MabSelect PrismA) showing dynamic binding capacity (DBC) as a function of cycle number using 0.3 M and 1.0 M NaOH sanitization solution every cycle
Cleaning and sanitization cycling (0.3 M NaOH, no protein load) on the prototype (MabSelect PrismA) resin showed no appreciable decrease in DBC (Figure 7b), whereas the same cycling on MabSelect SuRe showed a 17% decrease (Figure 7a). Cleaning and sanitization cycling (1.0 M NaOH, no protein load) on the prototype (MabSelect PrismA) resin showed a 28% decrease in DBC (Figure 7b), whereas the same cycling on MabSelect SuRe showed a 62% decrease (Figure 7a). Total caustic exposure (0.3 M or 1.0 M NaOH + 25mM NaOH strip) over 150 cycles was 37.5 h.
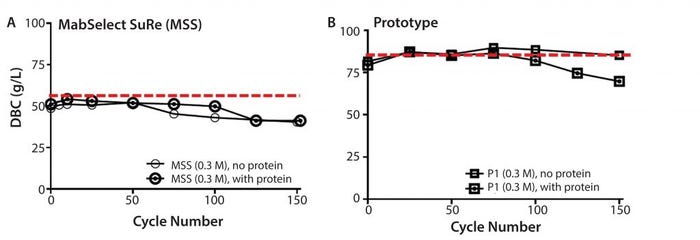
Figure 8: Resin lifetime study comparison between (left) MabSelect SuRe (MSS) resin (with protein load step) and (right) prototype (P1) (MabSelect PrismA) showing dynamic binding capacity as a function of cycle number using 0.3 M sanitization solution every cycle
Overall resin stability was further assessed by running full protein load cycles at 80% DBC with cleaning and sanitization steps performed on every cycle. Although sanitization (0.3 M NaOH, no protein load) on the prototype (MabSelect PrismA) resin showed little decrease in DBC, the same cycling (0.3 M NaOH, with protein load) showed approximately 15% decrease. The DBC decrease suggests protein loading further degrades the prototype resin beyond caustic only cycling and a typical DBC safety factor would be required to maintain high % yield (Figure 8b). Sanitization (0.3 M NaOH) on MabSelect SuRe showed no difference between protein and no protein load and showed ∼20% decrease (Figure 8a). The prototype appears to have higher caustic only stability with a total caustic exposure (0.3 M NaOH + 25mM NaOH strip) over 150 cycles of 37.5 h.
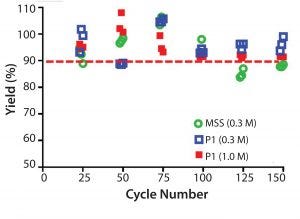
Figure 9: Resin lifetime study comparison between prototype (P1) (MabSelect PrismA) and MabSelect SuRe (MSS) resin (with protein load step) showing yield (%) as a function of cycle number using 0.3 M or 1.0 M NaOH sanitization
solution every cycle
Recovery values (% yield) for the prototype (MabSelect PrismA) resin were >90% over the duration of the resin lifetime study at 67 g MAb/L resin loading (80% of DBC at 4 minute residence time) (Figure 9). Recovery values (% yield) for MabSelect SuRe resin fell <90% after 100 cycles at 36 g MAb/L resin loading (80% of DBC at 4 minute residence time). Total caustic exposure (0.3 M NaOH + 25mM NaOH strip) over 100 cycles was 25 h.
Protein carryover was monitored by collecting the elution fraction of postsanitization (0.3 M and 1.0 M NaOH) blank runs after cycle 75 (every 25 cycles). All elution fractions were below one-thousandths of the previous elution pool concentration. Protein carryover was below the limit of quantitation (LOQ) for 1.0 M NaOH and slightly above for 0.3 M NaOH (Figure 10).
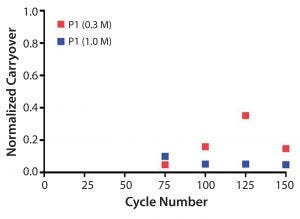
Figure 10: Normalized protein carryover of blank elution fraction after protein load and sanitization using 0.3 M NaOH or 1.0 M NaOH.
Leached protein A was monitored throughout the duration of the cycling studies performed with protein load. No significant changes were observed after cycle 25, and the relative levels of residual protein A leachate for the prototype (MabSelect PrismA) were about 4× higher than observed for the same conditions on MabSelect SuRe (Figure 11).
Continuous chromatography systems and skids are now commercially available to allow for continuous high protein loading beyond protein breakthrough. Periodic counter-current chromatography (PCC) can be operated with three or four columns attached in series with any two being loaded simultaneously. Previous findings suggest that batch protein A operations using short columns (bed height 5–10 cm) at relatively low loadings (<40 g MAb/L resin) could be a productive alternative (8–12 g/L/h) to continuous chromatography (3).
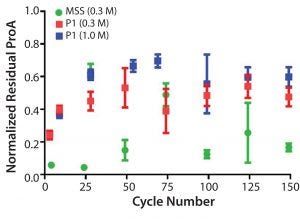
Figure 11: Normalized residual protein A leachate monitored in the elution pool as a function of cycle number
However, when using three or four column PCC with the prototype (MabSelct PrismA) resin at high loading capacity (>60 g MAb/L resin) combined with reduced residence time and reduced resin volume, downstream productivity could be improved by more than twofold. Depending on the optimization parameter goal (maximum productivity or maximum capacity), productivity values can reach 26 g/Lr/h (Lr = Lresin) (Table 1).
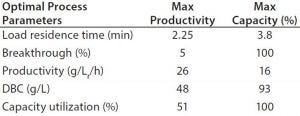
Table 1: GE Healthcare AKTA PCC Unicorn software productivity calculator using full breakthrough curves of prototype (MabSelect PrismA) resin (3 minute residence time).
Dynamic binding capacity for the prototype (MabSelect PrismA) protein A resin was observed to be >70 g MAb/L resin (3 minute residence time). Cycling studies, both with and without protein, indicate that the prototype (MabSelect PrismA) resin was significantly more caustic stable compared with its predecessor MabSelect SuRe resin. Average yields were >90% over the duration of resin lifetime studies. Total carryover was higher for the prototype (MabSelect PrismA) resin but well under the allowable carryover limit of one one-thousandths of average pool concentration from the previous cycle. The higher caustic stability of the prototype (MabSelect PrismA) resin allows not just more aggressive sanitization procedures, but also more flexible sanitization procedures — giving a wide range of options for specific applications. Using the prototype (MabSelect PrismA) resin in PCC mode could improve productivity by more than twofold.
Learn more: 
1 Tosoh BioScience Application Note A1528A: Protein A Chromatography — The Process Economics Driver in MAb Manufacturing.
2 Kobayashi S, Ueda Y. Comparing Protein A Resins for Monoclonal Antibody Purification. BioPharm Int. 26(12) 2013.
3 Kaltenbrunner O, et al. Continuous Bind-and-Elute Protein A Capture Chromatography: Optimization Under Process Scale Column Constraints and Comparison to Batch Operation. Biotechnol. Progress 32(4) 2016: 938–948; doi:10.1002/btpr.2291.
Bryan Dransart is a Senior Research Scientist I at Gilead Sciences. April Wheeler was formerly an Associate Scientist at Gilead Sciences (currently Technology Manager at MilliporeSigma). Tony Hong was formerly a Rearch Scientist 1 at Gilead (now retired). Chi Tran is Senior Research Associate II. Rafael Abalos is a Senior Research Associate II; Andrew Quezada is a Senior Research Associate I; ShiYu Wang is a Senior Research Associate I; Brian Kluck is a Research Scientist 1; and Nooshafarin Sanaie is a Senior Research Scientist II, all at Gilead Sciences.
You May Also Like