Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
https://www.alamy.com
At BPI Europe in April 2019, Dirk Gewert (business unit leader of bioproduction at Horizon Discovery) told BioProcess Insider that Chinese hamster ovary (CHO) cell lines haven’t changed much in the 30 years since they were first used in biomanufacturing (1). Only a few companies offered commercially available, production-ready CHO cell lines for large-scale manufacturing of biotherapeutics. “In all cases,” he said, “cell lines were selected by identifying high-expressing clones and focusing on process optimization to improve expression and other critical quality attributes.”
Gewart’s company launched a consortium of suppliers to improve that situation. Its key objective was “to leverage modern gene-editing tools to improve the biology at the heart of the cell, ultimately to generate a fundamentally improved CHO expression platform available to the entire industry for the manufacturing of routine biotherapeutics.” Using genome-editing tools such as clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9), the company is developing “a suite of engineered cell lines optimized with different attributes for more challenging molecules.”
By 2020, industry consultant Dan Piestun could report on impressive progress — both within and beyond the CHO consortium’s — in the pages of BPI (2). He described how gene editing is being used across the biopharmaceutical industry for
• therapeutic protein expression regulation with smart promoters and epigenomic reprogramming.
• epigenetic control of therapeutic proteins.
• protein folding and secretion of recombinant cells.
• control of posttranslational modifications.
And just this year, a team of authors from Merck expanded on the latter application (3). They described a number of successful applications of genetic engineering methods for control of both N- and O-glycosylation as well as tyrosine sulfation and other modifications in mammalian cells.
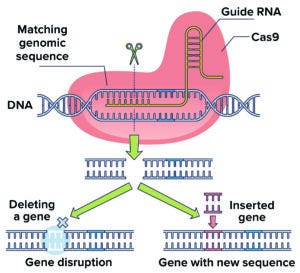
Figure 1: Adapted from a bacterial defense mechanism against virus infection, CRISPR/Cas9 technology uses enzymes and guide RNA to excise a target DNA sequence.
Industry researchers have gone beyond scratching the surface of what CRISPR/Cas9 (Figure 1) and other methods can do for biotherapeutic development and production. In a March 2022 talk at BPI West in San Diego, Ben Haley of Genentech (Roche) presented inspiring examples that lit the way toward even more potential uses (4).
Haley is a senior principal scientist in the molecular biology department at Genentech in South San Francisco, CA, where he’s worked for 12 years now. Previously, he was a postdoctoral fellow at the University of California (UC) at Berkeley, and he has also served as a visiting instructor at both California State University—East Bay and the UC Santa Cruz Extension school, where he taught on the principles of genetics and experimental methods in molecular biology, respectively. He earned a bachelor of science in biochemistry from the University of Massachusetts (UMass) at Amherst in 2000 and a doctorate in biomedical sciences and biochemistry from UMass Chan Medical School in 2005. Haley has published extensively on both cell biology and biomedicine.
BPI West Presentation
Haley’s presentation at BPI West was titled “End-to-End Platform Optimization for Loss-, Gain-, and Modification-of-Function Genetic Engineering.” In it, he defined genetic engineering as “technologies for genetic manipulation of cell-based systems, delivery/expression of the tools for genetic expression, and development of physiologically relevant cellular models.” He touted the importance of CRISPR/Cas9 technology as a powerful breakthrough with many applications. And he showed how the “knock-out” technique for loss-of-function screening has been the most widely used so far, with many researchers increasingly working with gain- and modification-of-function options as well.
“The rapid evolution of gene-editing tools has made it possible to suppress, activate, and mutate genomes at unprecedented scale and precision,” he stated in his presentation abstract. “However, the full potential for these technologies has yet to be realized, owing to differences in cell type or gene-dependent efficiency.”
Haley illustrated the utility of functional genomics, especially for drug discovery and developability assessments. For example, he showed how modification-of-function can be used to elucidate protein structure–function relationships and enhance recombinant therapies through in situ mutagenesis, as well as for evaluation of how such drugs affect tumors. Lineage tracing is helping scientists parse out biological pathways, especially related to tumor evolution. And both gain-of-function and loss-of-function work are helping to identify new targets for protein, DNA/RNA, and even cell therapies.
Our Conversation
Before BPI West, I chatted with Haley virtually about the subject of his talk there.
What does your team do at Genentech? My team develops molecular genetic tools to support a range of basic and clinical research programs.
Over a decade ago, zinc-finger nucleases (ZFNs) were touted as an exciting technology for genetic engineering. And recent approaches to structural diversification for their increased precision seem to have revitalized interest in them. What are their general drawbacks and benefits for cell-line engineering? ZFN technologies largely helped set the stage for the current era of genome modification tools. However, a primary downside of ZFNs comes from the level of expertise needed to design, clone, and express functional, specific nucleases for editing your desired target gene. Although it would be unwise to recommend the use of ZFNs over CRISPR/Cas for the average researcher, some forms of ZF-based modifiers could have niche applications. In particular, Kruppel-associated box zinc-finger fusion proteins (KRAB-ZFPs) have shown promise as gene-silencing tools. They have the advantage of needing only a single, relatively compact protein for activity compared with the larger, more complex CRISPR-based technologies.
Transcription activator-like effector nucleases (TALENs) received a lot of attention 10 years ago, but I haven’t come across many reports about their use in our industry since then. Is anyone using this approach in cell-line engineering for biotherapeutic production? Similar to ZFNs, TALENs require unique know-how to design effective genome editors. However, they may provide access to parts of the genome that are otherwise difficult to target because of sequence limitations inherent to ZFN or CRISPR-based methods.
What makes CRISPR so much more exciting than those other technologies? I would say that CRISPR “democratized” genome editing. It made the process of gene manipulation accessible to virtually anyone who works with cells (in a dish or in an organism) as part of their research. This comes from the high functional activity of Cas proteins combined with the simplicity of designing a new, short-guide RNA sequence for each desired target gene — rather than an entirely new protein/dimer as would be the case with ZFNs or TALENs. CRISPR is also highly scalable thanks to its small-RNA–guided nature, so genome-wide targeting libraries can be created and applied for functional genomic screens.
Do viral-vectored gene-transfer methods or phage-based techniques have potential for use in cell-line engineering for biopharmaceutical production? For certain. Viral vector technologies are critical for cell-line development in a basic research setting. This does become complicated when cells are being developed for clinical or manufacturing purposes, but fortunately there are several nonviral, transient gene-editing methods available these days — including Cas9-ribonucleoproteins (RNPs), mRNA-based delivery of gene editors, and so on.
What are some other important platform technologies in cell-line engineering? I can’t think of a better time than now to be interested in cell-line engineering technology. So many different applications are maturing or being made available every day. In particular, I’m excited about the potential for next-generation “genetic switches” that use a combination of chemical and synthetic biology so that we can toggle gene expression on or off.
I think of functional genomics as being more of a “drug-discovery thing.” Can you explain its utility in cell-line development? Functional genomics covers a lot of ground, and it has a number of applications for cell-line development. For example, researchers can activate or knock out every gene in their favorite cell type’s genome, providing the opportunity to identify factors that previously had been unexplored when it comes to cell viability, nutrient sensing, protein output, and so on. There are also “passive” approaches to functional genomics that make use of next-generation lineage-tracing tools to offer insight into clonal variation and stability.
At BPI West you focused on optimizing these types of platforms. Can you describe the general approach to doing so? Because my laboratory works across therapeutic areas, we take great pains to ensure that the technologies we develop can be applied across many cell types. This is an extreme challenge for one group because we can’t be experts in all cellular contexts, but Genentech provides a remarkably collaborative environment. As such, we work directly with many other colleagues to identify and optimize their prioritized cell lines for single gene manipulation or genome-scale screens.
Timelines are increasingly important in cell-line development. Automation technologies have helped many companies speed up their engineering and selection processes. Have you taken advantage of such instrumentation in your work? To date, most of our cell-line engineering work has come through manual handling. Yet we do recognize the value and potential for automation, so we’ve been investing recently in automated imaging and single-cell cloning systems.
In your talk, you mentioned differences in cell type can affect the complexity of genetic-engineering platform development. What are some examples? Both the cell type of interest and the desired gene-editing outcome affect everything about the approach you might take for genetic engineering. This includes the reagent delivery method, the specificity or potency of your genome-modifying tools, and the way in which your cells react to the newly edited gene in subsequent bulk populations or single-cell cultures.
You mentioned that differences in gene-specific efficiencies as another complication. How do those generally manifest? Gene expression strength and isoform use can vary widely across cell types (and sometimes within otherwise homogeneous populations). Having deep knowledge of these patterns is critical toward understanding the most effective gene-targeting approach and whether or not a gene-editing process has created the desired molecular or phenotypic outcome.
Participants |
References
1 Stanton D. CRISPR Consortium Launched to Improve CHO Cell Lines. BioProcess Insider 17 April 2019; https://bioprocessintl.com/bioprocess-insider/upstream-downstream-processing/crispr-consortium-launched-to-improve-cho-cell-lines.
2 Piestun D. Use of CRISPR and Other Gene-Editing Tools in Cell Line Development and Engineering. BioProcess Int. 18(9) 2020: i8–i12; https://bioprocessintl.com/analytical/cell-line-development/use-of-crispr-cas9-and-other-gene-editing-tools-in-cell-line-development-and-engineering.
3 Zhang Y, Du Z, Liu R. Posttranslational Modifications and Their Control in CHO Culture. BioProcess Int. 20(1–2) 2022: 16–20; https://bioprocessintl.com/upstream-processing/expression-platforms/posttranslational-modifications-and-their-control-in-cho-culture.
4 Haley B. End-to-End Platform Optimization for Loss-, Gain-, and Modification-of-Function Genetic Engineering. BioProcess International US West Conference and Exhibition: San Diego, CA, 15 March 2022.
Further Reading
Cuellar TL, et al. Application of CRISPR for Pooled, Vector-Based Functional Genomic Screening in Mammalian Cell Lines. Genome Editing and Engineering: From TALENs, ZFNs and CRISPRs to Molecular Surgery. Cambridge University Press: Cambridge, UK, August 2018; 209–222.
Dangi AK, et al. Cell Line Techniques and Gene Editing Tools for Antibody Production: A Review. Front. Pharmacol. 12 June 2018; https://doi.org/10.3389/fphar.2018.00630.
Ley D, et al. Reprogramming AA catabolism in CHO cells with CRISPR/Cas9 genome editing improves cell growth and reduces byproduct secretion. Metabol. Eng. 56, December 2019: 120–129: https://doi.org/10.1016/j.ymben.2019.09.005.
Sargeva D, et al. CRISPR/Cas9 as a Genome Editing Tool for Targeted Gene Integration in CHO Cells. Meth. Mol. Biol. 1961, 2019: 213–232; https://doi.org/10.1007/978-1-4939-9170-9_13.
Senger K, Haley B. Nonviral Alternatives to Delivery of CRISPR-Cas Gene Editing Components. CRISPR-/Cas9 Based Genome Editing for Treating Genetic Disorders and Diseases. CRC Press: Boca Raton, FL, January 2022; 183–223.
Cheryl Scott is cofounder and senior technical editor of BioProcess International, part of Informa Connect; 1-212-600-3429; [email protected].
You May Also Like