Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
Transmission electron micrograph of a mesenchymal stromal cell ROBERT M. HUNT (HTTPS://EN.WIKIPEDIA.ORG)
Mesenchymal stromal cells (MSCs) are multipotent, self-renewing progenitor cells that can differentiate into adipocytes, chondrocytes, and osteocytes (1). Cultured MSCs are plastic-adherent and spindle-shaped, and they express cell-surface markers CD44, CD73, CD90, and CD105, but not CD14, CD34, CD45, CD11b, CD79a, CD19, or HLA-DR (2, 3). First isolated from bone marrow (BM), human MSCs have been investigated extensively in clinical studies. MSCs also have been isolated from adipose tissue (4) and peripheral blood (5). Perinatal organs and tissues such as amniotic membrane, placenta, and umbilical cord (UC) also have been shown to be rich sources of MSCs (6). The UC consists of a cord lining (CL), Wharton’s jelly (WJ), and a perivascular region, all of which have been documented as sources for isolation and expansion of MSCs (7–9). WJ reportedly yields 10,000 to 4.7 million MSCs per centimeter of UC before culture; CL reportedly yields 125 million per centimeter of UC at first passage (8, 9). That makes UCs a readily available source of MSCs that poses no ethical issues because they often are considered to be medical waste after childbirth (10).
Several groups have attempted to isolate and define MSCs from different compartments of the UC (10–13). Notably, Subramanian et al. attempted to extract MSCs from specific anatomical regions of noncryopreserved UC (11): amniotic membrane, subamniotic layer, WJ, and perivascular regions. As in many earlier studies, the authors faced difficulties in identifying and separating each region of the UC. Here, we compare two main regions of the UC for the sake of simplicity: CL consisting of amniotic and subamniotic layers, and WJ including the perivascular region.
Manufacture of MSC-based cell-therapy products is guided by the US Food and Drug Administration (FDA) rules for human cells, tissues, and cellular and tissue-based products (HCT/Ps) and by European Medicines Agency (EMA) regulations for advanced therapy medicinal products (ATMPs), under which minimally manipulated tissue/cell sources are recommended. To that end, MSCs can be cryopreserved in raw UC tissue and then thawed at a later time for extraction and/or further expansion to use in prospective autologous transplants. Nonenzymatic methods have been used successfully for high-yield isolation of UC MSCs from cryopreserved UC (14). During comparison studies on MSC extraction from cryopreserved UC by explant and enzymatic dissociation methods, we have found that the enzymatic dissociation method provides higher yields of viable WJ-MSCs (15).
Because of their low immunogenicity (16), human MSCs increasingly are used in allogeneic cell therapy applications (17). Dosages of MSC-based cellular therapies range from 1–2 million cells per kilogram of patient body weight, depending on the condition being treated (18, 19). Such therapies would require scaling up of MSC expansion in a current good manufacturing practice (CGMP)–compliant environment using optimized culture systems. Current large-scale MSC expansion methods typically use culture medium that is supplemented with a common concentration (10%) of fetal bovine serum (FBS) (20), which could introduce inadvertent variability to the manufactured cell product.
By contrast, serum-reduced media such as PTT-6 culture medium developed by CellResearch Corp (CRC) in Singapore, can reduce or eliminate variability in the manufacturing of MSCs. This medium is used for production of CorLiCyte CL-MSCs in an ongoing phase 1 clinical trial for treatment of chronic diabetic foot ulcers at the University of Colorado’s Anschutz Medical Campus (21).
Here, we define and compare MSCs isolated from two main UC regions: the CL and WJ. We used cryopreserved UC tissues to verify that extracted MSCs are viable after cryopreservation (postthaw). Consequently, we determine whether the PTT-6 culture medium would alter characteristics of UC-MSCs after repeated passaging and culturing. We also investigate the extraction efficiency of two methods coupled with the two culture media used to isolate MSC populations from CL and WJ thawed after cryopreservation in UC tissue (Figure 1).
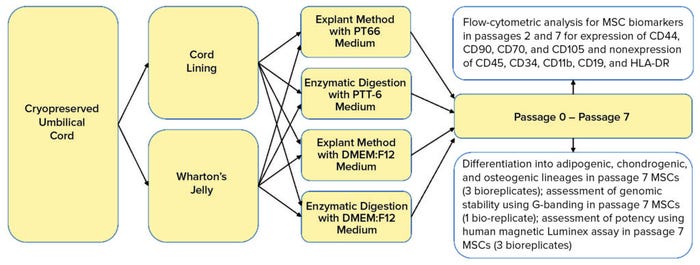
Figure 1: Umbilical cord lining and Wharton’s jelly were separated carefully from cryopreserved umbilical cord soaked in wash buffer. Tissues were explanted or digested and cultured in either PTT-6 medium or DMEM:F12 medium for six passages. Some cells were frozen at passages 1 and 6 for flow-cytometric analysis; others were assessed at passage 7 for genomic stability and therapeutic potential using differentiation assays, G-banding, and a human magnetic Luminex assay.
Materials and Methods
MSC Isolation, Culturing, and Population Doubling Time (PDT): Six UCs were collected (with informed consent) from both vaginal and Caesarean-section deliveries, with gestational ages ranging 37–42 weeks and UC lengths >10 cm. We excluded UCs that were microbially contaminated or found to have viral presence: e.g., human immunodeficiency virus (HIV), hepatitis B virus (HBV), or hepatitis C virus (HCV). The UC samples were transported dry in sterile polyethylene canisters and washed before use. Final concentrations of penicillin, streptomycin, and amphotericin B were 10,000 units/mL, 10 mg/mL, and 25 µg/mL respectively.
Next, the samples were sliced into 0.5-cm thick discs using a scalpel and rinsed repeatedly with wash buffer to remove residual blood. UC discs were transferred into sterile cryovials and immersed in cryoprotectant composed of sterile HyClone Dulbecco’s modified Eagle’s medium: Nutrient Mix F-12 (DMEM:F12) from Cytiva and 10% dimethyl sulfoxide (DMSO) from WAK-Chemie. Samples then were subjected to controlled-rate freezing of –1 °C/minute down to –40 °C, when they were brought down to –90 °C at –10 °C per minute before subsequent storage in vapor-phase liquid nitrogen tanks. All samples were frozen 24–48 hours from the point of collection and then stored at –196 °C for at least seven days before being thawed in a water bath at 37 °C for about a minute.
Thawed UC discs were transferred to a wash solution containing 2% v/v antibiotic and antimycotic for five minutes, then soaked in the above-described wash buffer for another five minutes. Subsequently, all UC discs were sliced in half and soaked in one of the following with a 1% v/v antibiotic and antimycotic solution:
• proprietary PTT-6 culture medium comprising DMEM:F12 from Lonza, medium 171 (M171) from Life Technologies (Thermo Fisher Scientific), 0.2% epidermal growth factor (EGF), 0.035% insulin, and 2.5% v/v FBS from Phoenix Scientific (22)
• DMEM:F12 (with 2.5 mM l-glutamine) complete growth medium supplemented with 10% v/v FBS.
We kept the soaked samples at 37 °C in a humidified 5% CO2 incubator for three to four days, after which the WJ could be distinguished easily from the CL by their visible difference in texture. Without transferring the growth medium, we laid out the halved UC discs on a petri dish and carefully separated those components with a scalpel.
For the explant method, separated CL and WJ tissue from each medium were diced into 2-mm2 to 5-mm2 pieces and spotted onto 10-cm2 culture dishes from Corning. We left those to dry for 10 minutes for cell adherence, then added 5 mL of culture media to each dish and incubated them for three hours before adding another 5 mL of culture media to cover the tissue pieces.
For the enzymatic digestion method, separated CL and WJ were diced and placed in a proprietary blend of enzymes (15), then incubated with shaking at 37 �°C for two hours. After incubation, the mixtures were centrifuged at 500g for five minutes, then washed twice with their respective culture media. Then we seeded those mixtures into 10-cm culture dishes with 10 mL of culture medium.
All cultures were kept at 37 °C in a humidified 5% CO2 incubator with a media refresh on the seventh day. Thereafter, we monitored the cultures for growth, refreshing the media every three to four days. Whenever a culture reached ~90% confluency, we took an image of its cell morphology and then passaged the cells as follows.
Spent culture media was aspirated completely, and the cell monolayer was washed twice with Dulbecco’s phosphate-buffered saline (DPBS) from Biological Industries to remove nonadherent cells and debris. The MSCs were detached by incubation with 0.25% w/v trypsin solution from Life Technologies (Thermo Fisher Scientific) at 37 °C for five to 10 minutes. For postexperimental analysis, cultures from passages 1 and 6 (P1, P6) were treated with Accutase cell-detachment solution (Thermo Fisher Scientific) at 37 °C for 5–10 minutes until all cells were lifted. At the fifth passage (P5), we prepared triplicate subcultures to ensure that sufficient cells would be harvested in P6. Cells were enumerated on a hemocytometer with either a trypan blue exclusion assay or crystal violet nuclei staining. All cultures were subcultured at a seeding density of
3.75 × 105 viable cells per dish (~6,800 cells/cm2). We cryopreserved the released cells in their respective culture media supplemented with 10% v/v DMSO.
To determine the PDT, we used the formula PDT = t × ln(2)/ln(n2⁄n1), where n2 is the harvesting cell number, n1 is the plating cell number, and t is the number of hours taken to reach the next passage (23). We performed all statistical analysis with GraphPad Prism software (Dotmatics), with data presented as mean ± standard deviation (SD) and multiple t-tests used. For all data presented herein, the level of significance is either p < 0.05 or not significant (n.s.).
Flow Cytometry Analysis: To ascertain the purity of MSCs from different culture conditions, we restarted cultures with cryopreserved cells taken from P1 and P6. After three days of culturing those to P2 and P7, respectively, cells were detached using Accutase solution and stained with a human MSC analysis kit from BD Bioscience. Stained cells were fixed with fixation buffer (BD Bioscience) containing 4% w/v formaldehyde.
Using flow-cytometric analysis of six biological replicates, we assessed cells from each extraction and culture condition for expression of MSC biomarkers (CD44, CD73, CD90, and CD105) and for nonexpression of hematopoietic lineage cell markers (CD45, CD34, CD11b, CD19, and HLA-DR). We used FlowJo software from Tree Star for post-acquisition analysis (Figure 2). Using the gated percentages of double-positive CD90-FITC and CD44-PE biomarkers multiplied by double-positive CD105-PerCP-Cy5.5 and CD73-APC biomarkers (percentages from FlowJo), we calculated the quadruple-positive MSC percentages of each sample for all conditions at P2 and P7. Subsequently, to determine the purity of MSCs in each set of conditions at P2 and P7, we calculated the mean and SD of quadruple-positive MSC percentages for each.
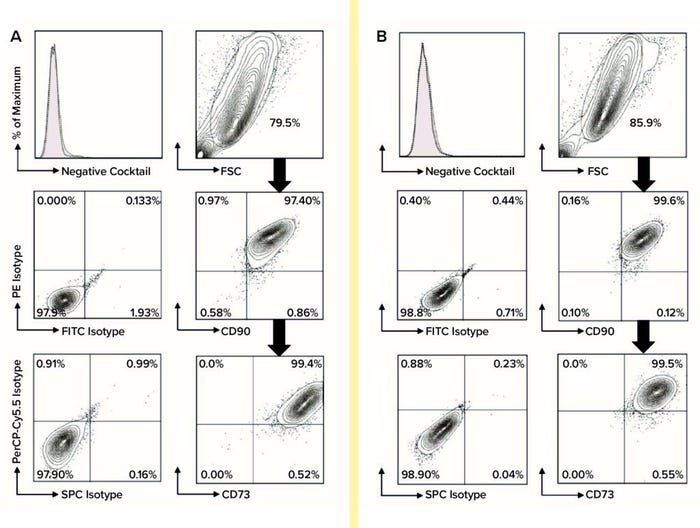
Figure 2: For flow-cytometric analysis of MSCs at passage 7, cells were stained using anti-CD73-APC, anti-CD90-FITC, and anti-CD105- PerCP-Cy5.5 along with relevant isotype controls using a Stemflow hMSCs analysis kit from BD Bioscience. Markers associated with hematopoietic lineage cells also were stained using a negative cocktail of PE-conjugated antibodies specific for CD45, CD34, CD11b, CD19, and HLA-DR. Controls are represented by the left column in each category, with MSC surface-marker expression in each right column. MSCs were gated in the FSC-SSC plot and subgated to reveal expression levels of CD44, CD73, CD90, and CD105; subgate cells mostly appear in the top-right quadrant of the plot, indicating positive biomarker expression. A total of six biological replicates were analyzed for each condition: (A) flow-cytometric analysis of DMEM:F12–cultured MSCs and (B) flow-cytometric analysis of PTT-6–cultured MSCs.
Differentiation Assay: To assess the “stemness” of repeatedly passaged MSCs, we induced P7 MSCs (n = 3 bioreplicates) into adipogenic, chondrogenic, and osteogenic lineages using StemPro differentiation kits from Life Technologies (Thermo Fisher Scientific). These cultures were restarted from cryopreserved cells at P6. After three days of recovery, cells were trypsinized and seeded into 12-well plates at 2 × 104 cells/well for cells cultured in both PTT-6 and DMEM:F12. Each differentiation assay had a duplicated 12-well plate to serve as a control.
Cells were maintained in their respective culture media for up to three days for differentiation plates and seven days for control plates. Differentiation-plate media were replaced with adipogenic, chondrogenic, and osteogenic differentiation media and then cultured for 21 days more. At the end of the culture, cells were fixed with fixation buffer containing 4% w/v formaldehyde.
We stained the resultant cultures for the presence of adipocytes, chondrocytes, and osteocytes using Oil Red O, Safranin O, and Alizarin Red S dyes, respectively, all from MilliporeSigma/Merck. Stained plates were observed under an inverted phase-contrast microscope using bright-field illumination, and we captured images using Infinity Analyze software from Lumenera.
Karyotyping Analysis: We used karyotyping analysis to assess gross chromosomal abnormalities of cells from repeated passaging. To cover all experimental conditions, we tested P7 MSCs (n = 1) from PTT-6–CL–explant and DMEM:F12–WJ–digest conditions. Samples were revived from cryopreservation and seeded into T-25 culture flasks (Corning), then cultured to P7 at 70–80% confluency. Next, those cells were subjected to human chromosomal G-banding karyotyping analysis at the Cytogenetics Laboratory of the KK Women’s and Children’s Hospital in Singapore. Results were returned as a cytogenetics report with an image of chromosomes in an orderly arrangement.
Potency Assay: To determine the therapeutic efficacy of MSCs from explanted tissues after repeated passages, we revived P7 MSCs (n = 3) from cryopreservation and cultured in their respective media to P7 cells from the following conditions: PTT-6–CL–explant, PTT-6–WJ–explant, DMEM:F12–CL–explant, and DMEM:F12–WJ–explant. We did not study enzyme-digested tissues because lingering enzymes in culture could affect the quantification levels of proteins secreted by MSCs.
After two days, we collected and cryopreserved supernatant from those cultures, then sent those samples to ClinImmune Labs at the University of Colorado–Denver for quantification of secreted proteins by a human magnetic Luminex assay (R&D Systems). Quantified cytokines from those supernatants included angiopoietin-1 (Ang-1), vascular endothelial growth factor (VEGF), platelet-derived growth factor consisting of the A homodimer (PDGF-AA), platelet-derived growth factor consisting of the B homodimer (PDGF-BB), hepatocyte growth factor (HGF), interleukin-10 (IL-10), basic fibroblast growth factor (bFGF), and transforming growth factor beta 1 (TGF-β1).
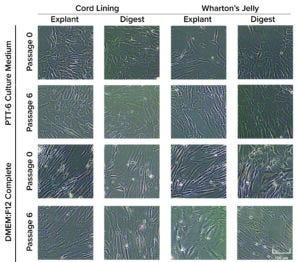
Figure 3: Morphological differences were observed in mesenchymal stromal cells (MSCs) cultured in DMEM:F12 medium after multiple passages, but no morphological differences were observed with MSCs cultured in PTT-6 medium. Six total biological replicates were analyzed from each set of conditions.
Results
Human MSCs were extracted from both CL and WJ using explant or digestion methods. Initial passage showed that MSCs cultured in both PTT-6 and DMEM:F12 media displayed fibroblasticity, spindle-shaped morphology, and adherence to plastic surfaces when maintained in standard culture conditions (37 °C, 5% CO2). At P6, cells cultured in PTT-6 medium maintained the same morphology observed in P0. However, MSCs cultured in DMEM:F12 (regardless of tissue source or extraction method used) showed enlarged and polygonal morphologies (Figure 3) after passage.
Next, to determine the proliferation rates for isolated MSCs under the investigated conditions, we plotted mean PDT graphs and compared the results (Figure 4, left). Cells cultured in PTT-6 medium showed significantly shorter PDTs (50.1 ± 9.7 hours) than those in DMEM:F12 (98.4 ± 37.5 hours). Note that the PDT of PTT-6–cultured cells remains stable, with much less fluctuation than for cells in DMEM:F12, making PTT-6 medium a better candidate for manufacturing process standardization. Cells cultured in that medium showed no significant difference in PDT regardless of tissue source or extraction method. This suggests that PDT depends more on the type of culture medium used than on either the extraction method or tissue source, with no significant differences observed (p > 0.05) when the same medium was used.
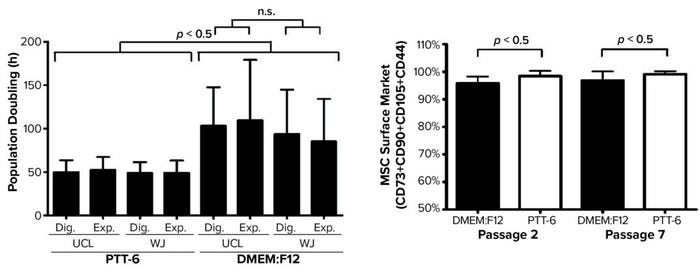
Figure 4: (LEFT) To compare mean ± SD population doubling time (PDT) across all experimented conditions, a total of six biological replicates were analyzed for each condition (Dig. = digest, Exp. = explant). (RIGHT) Purity profiles between passages 2 and 7 (P2, P7) reveal constant purity for all PTT-6–cultured MSCs after multiple passages; DMEM:F12–cultured MSCs displayed lower expression at P2, increasing by P7. Significant differences in cell-surface marker expression were seen for MSCs cultured in DMEM:F12 and PTT-6 at P2 and P7 (n.s. = not significant). Six total biological replicates were analyzed from each set of conditions.
Based on standards recommended by the International Society for Cell and Gene Therapy (ISCT), MSC purity is defined by positive expression of a group of specific surface markers —CD44, CD73, CD90, and CD105 — and a lack of expression for hematopoietic stem cell (HSC) surface markers CD34, CD45, CD11b, CD19, and HLA-DR. Decreasing expression levels of MSC-specific surface markers implies a cell purity reduction. So we compared the purity of MSCs cultured in different conditions across passages by examining the expression levels of MSC-specific markers (Figure 4, right). Tabulated flow-cytometry results revealed that PTT-6–cultured cells maintained persistent high purity across multiple passages (98.4 ± 1.9% at P2, 99.1 ± 1.1% at P7), whereas DMEM:F12–cultured cells expressed marginally lower levels of those markers at P2 (95.8 ± 2.5%) that increased by P7 (96.8 ± 3.3%).
When we compared tissue sources and extraction methods, we found no observable differences in MSC purity (data not shown), which indicates that it is medium dependent. No cultured cells expressed HSC surface markers. PTT-6–cultured cells demonstrated significantly higher surface-marker expression than did DMEM:F12–cultured cells (p < 0.05) at both P2 and P7 (Figure 4, right). On comparing the median fluorescence intensity (MFI) of individual MSC biomarkers from P2 and P7 (Figure 5), we found increased CD73 expression in DMEM:F12 cultured cells and an increase in the expression of CD90 in PTT-6–cultured cells.
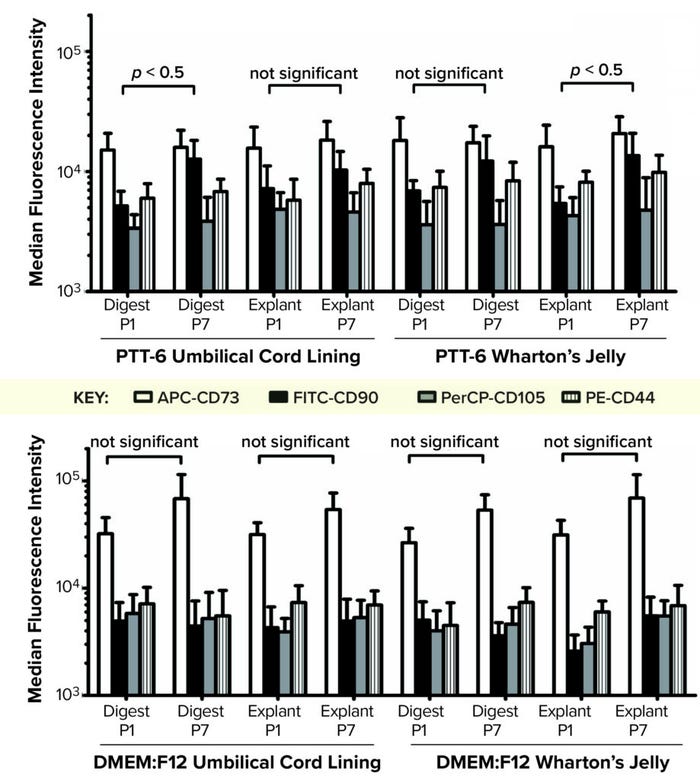
Figure 5: To determine the median fluorescence intensity (MFI) of MSCs in different culture conditions, MFIs for each MSC biomarker (CD73-APC, CD90-FITC, CD105-PerCP, and CD44-PE) were plotted for each set of conditions at passages 1 and 7 (P1, P7). (TOP) Marginal increase of CD90-FITC can be seen from P1 to P7 for all PTT-6 cultures. (BOTTOM) No significant increase of CD73-APC is seen for the DMEM:F12 conditions. A total of six biological replicates were analyzed from each set of conditions.
To determine their differentiation potential, we subjected MSCs from all experimental conditions at P7 to trilineage differentiation assays. Results showed that all MSCs were able to differentiate into adipogenic, chondrogenic, and osteogenic lineages (Figure 6, top).
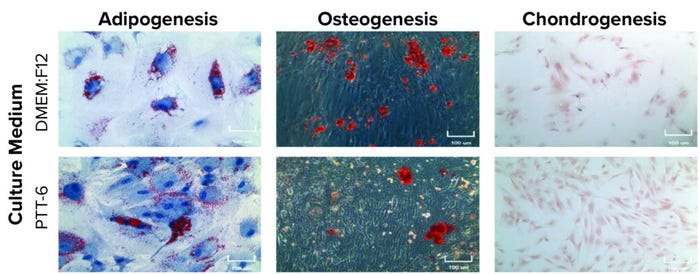
Figure 6: Potency and genetic stability of cultured MSCs; (TOP) trilineage differentiation of MSCs cultured in PTT-6 and DMEM:F12 media at passage 7 (P7), 100-μm scale; (BOTTOM) karyotyping analysis of P7 cells cultured as indicated, with no gross chromosomal abnormalities observed, suggesting genomic stability (a single biological replicate was analyzed for each condition).
Karyotyping analysis was applied to examine the genetic stability of the cultured cells. No obvious chromosomal abnormalities were found on P7 cells cultured in any set of conditions (Figure 6, bottom), suggesting that all MSCs cultured in this study were stable genetically after multiple passages regardless of tissue source, extraction method, or medium used.
To study potency and therapeutic potential of our MSCs following multiple passages, we used a cytokine secretion assay. Results showed significantly higher secretion levels (p < 0.05) of IL-10, VEGF, PDGF-AA, Ang-1, HGF, and TGF-β1 in PTT-6–cultured MSCs than in DMEM:F12–cultured cells. By contrast, DMEM:F12–cultured MSCs showed significantly higher secretion levels (p < 0.0001) for bFGF. No PDGF-BB secretion was detected in any experimental conditions (Figure 7).
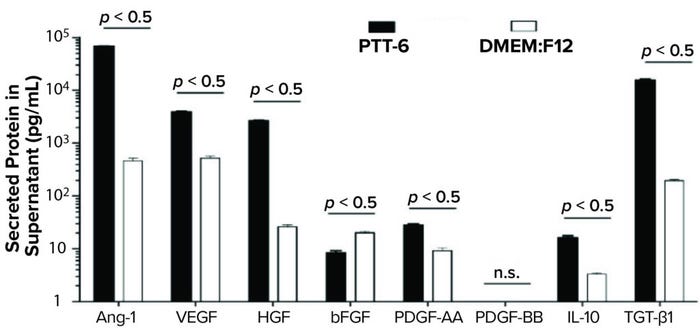
Figure 7: Secreted proteins from cells cultured as indicated and quantified by human magnetic Luminex assay. PTT-6 MSCs showed significantly higher levels of IL-10 (16.1 ± 1.6 pg/mL), VEGF (3,898 ± 171.9 pg/mL), PDGF-AA (28.2 ± 1.4 pg/mL), Ang-1 (67,653 ± 1,261.2 pg/mL), HGF (2647 ± 101.7 pg/mL), and TGF-β1 (15,590.3 ± 486.3 pg/mL) than those of MSCs cultured in DMEM:F12, which were 3.3 ± 0.2 pg/mL for IL-10, 521 ± 47.7 pg/mL for VEGF, 20.2 ± 1.3 pg/mL for bFGF, 9.2 ± 1.2 pg/mL for PDGF-AA, 465 ± 57.6 pg/mL for Ang-1, 26 ± 2.4 pg/mL for HGF, and 195.5 ± 8.4 pg/mL for TGF-β1. The latter group secreted significantly higher levels of bFGF (20.2 ± 1.3 pg/mL) than those cultured in PTT-6 (8.4 ± 0.8 pg/mL). No PDGF-BB secretion was detected in either group, and a total of three biological replicates were analyzed for each set of conditions.
Discussion
UCs provide a rich reservoir of MSCs that can be harvested and expanded for clinical use (8, 9). The intrinsic characteristics of UC-MSCs could be affected adversely by extraction using enzymatic digestion followed by long-term culture in FBS-supplemented medium. Furthermore, using a common concentration (10%) of animal-based serum supplements can introduce variability to manufactured MSCs (20). Our goal here was to compare the characteristics and therapeutic potential of MSCs derived from cryopreserved CL and WJ using different extraction methods (explant and enzymatic digestion) and culture media (CRC-patented PTT-6 culture media and a commercially available DMEM:F12 medium supplemented with 10% FBS).
Large-scale expansion of MSCs for therapeutic purposes requires them to proliferate efficiently within a short time while preserving their desirable stem-cell qualities. So we assessed cell morphology, proliferative capacity, purity, differentiation potential, and human chromosomal karyotyping to measure differences between the extraction techniques and culture medium formulations. Overall, the PDT of PTT-6–cultured MSCs was significantly shorter than that of their DMEM:F12–cultured counterparts, indicating a shorter time required to reach desired cell populations. Moreover, the highly variable PDTs found in all DMEM:F12–cultured MSCs could be related to variability in the constituents of FBS. Because shorter and more stable PDTs are desirable, we deem PPT-6 medium to be arguably superior for MSCs culture. Morphological changes have been reported in other studies of MSCs cultured in FBS-supplemented media over time (24).
PTT-6–cultured cells consistently maintained high surface marker expression levels following multiple passages. In fact, we found that expression of the cell surface marker CD90 expression marginally increased from P2 to P7 across all conditions in PTT-6 culture medium. That increased expression might help maintain the undifferentiated state of MSCs: Reduction of CD90 has been shown to regulate the transition of MSCs toward differentiation (25). By contrast, expression of CD73 increased across all conditions cultured in DMEM:F12. That could signal a differentiation pathway toward either chondrogenic or osteogenic differentiation (26). Comparing the different culture media showed that all conditions fulfilled the multipotent-differentiation criteria by demonstrating the capacity for trilineage differentiation to adipocyte, osteoblast, and chondroblast cell lines (27).
Use of MSCs for treating wounds with chronic inflammation has been studied clinically (28, 29). Critical proangiogenic factors (e.g., Ang-1, VEGF, HGF, bFGF, PDGF-AA, and PDGF-BB) and antiinflammatory cytokines (e.g., IL-10 and TGF-β1) have been identified as playing important roles in the acute inflammatory, proliferative, and remodeling phases of wound healing (30). Moreover, the ability of MSCs to secrete high levels of proangiogenic and antiinflammatory cytokines has been shown to enhance wound healing (31). Here, we have observed significantly higher secretion levels of Ang-1, VEGF, HGF, PDGF-AA, IL-10, and TGF-β1 from PTT-6–cultured MSCs than from DMEM:F12–cultured MSCs, which had significantly higher secretion levels of bFGF. Negligible secretion levels of PDGF-BB were quantified in all experimental conditions.
The lower secretion levels of bFGF and PDGF-BB in our results are in agreement with a previous study suggesting that neither of those angiogenic factor plays a key role in angiogenesis during wound healing (32). We included our list of proangiogenic factors as part of CGMP manufacturing potency control, which has been vetted and approved by the US FDA for chemistry, manufacturing, and controls (CMC).
Medium of Choice
Our results show that CellResearch Corporation’s patented PTT-6 culture medium significantly hastens UC-MSC expansion from cryopreserved CL and WJ, as compared with conventional DMEM:F12 culture media, while maintaining MSC “stemness,” trilineage differentiation potential, and chromosomal stability. Such validated quality fulfills minimal ISCT criteria, and genetic stability supports the potential of these cells for clinical therapeutic use. Enhanced secretion of proangiogenic and antiinflammatory cytokines from the PTT-6–cultured MSCs further supports the suitability of that medium in ex vivo expansion of UC-MSCs for clinical treatment of wounds with chronic inflammation. Further animal-based studies are warranted to investigate the suitability of using PTT-6–expanded UC-MSCs for other treatments such as for chronic wounds.
Acknowledgments
We are grateful to Mary Gan and Cheong Kum Foong (KK Women’s and Children Hospital cytogenetics laboratory) for their help in the human chromosomal G-banding karyotype analysis and to Brian Freed and Predrag Šerbedžija (ClinImmune Labs) for their help in quantification of secreted proteins using the human magnetic Luminex assay. Part of this work was presented as an abstract during the International Society of Cellular Therapy (ISCT) 2019 Annual Meeting in Melbourne, Australia. This study was funded by Cordlife Technologies Pte. Ltd (a subsidiary of Cordlife Group Limited) and CellResearch Corporation Pte. Ltd., of which the authors are employees.
References
1 Nardi NB, da Silva Meirelles L. Mesenchymal Stem Cells: Isolation, In Vitro Expansion and Characterization. Stem Cells 2008: 249–282; https://doi.org/10.1007/3-540-31265-X_11.
2 Van Pham P, et al. Isolation and Proliferation of Umbilical Cord Tissue Derived Mesenchymal Stem Cells for Clinical Applications. Cell Tiss. Banking 17(2) 2016: 289–302; https://doi.org/10.1007/s10561-015-9541-6.
3 Majore I, et al. Identification of Subpopulations in Mesenchymal Stem Cell-Like Cultures from Human Umbilical Cord. Cell Commun. Sign. 7(1) 2009: 1–8; https://doi.org/10.1186/1478-811x-7-6.
4 Bunnell BA, et al. Adipose-Derived Stem Cells: Isolation, Expansion and Differentiation. Methods 45(2) 2008: 115–120; https://doi.org/10.1016/j.ymeth.2008.03.006.
5 Chong PP, et al. Human Peripheral Blood Derived Mesenchymal Stem Cells Demonstrate Similar Characteristics and Chondrogenic Differentiation Potential to Bone Marrow Derived Mesenchymal Stem Cells. J. Orth. Res. 30(4) 2012: 634–642; https://doi.org/10.1002/jor.21556.
6 Marquez-Curtis LA, et al. Mesenchymal Stromal Cells Derived from Various Tissues: Biological, Clinical and Cryopreservation Aspects. Cryobiology 71(2) 2015: 181–197; https://doi.org/10.1016/j.cryobiol.2015.07.003.
7 Nagamura-Inoue T, He H. Umbilical Cord-Derived Mesenchymal Stem Cells: Their Advantages and Potential Clinical Utility. World J. Stem Cells 6(2) 2014: 195; https://doi.org/10.4252/wjsc.v6.i2.195.
8 Lim IJ, Phan TT. Epithelial and Mesenchymal Stem Cells from the Umbilical Cord Lining Membrane. Cell Transplant. 23(4–5) 2014: 497–503; https://doi.org/10.3727/096368914×678346.
9 Vangsness Jr. CT, Sternberg H, Harris L. Umbilical Cord Tissue Offers the Greatest Number of Harvestable Mesenchymal Stem Cells for Research and Clinical Application: A Literature Review of Different Harvest Sites. Arthroscopy 31(9) 2015: 1836–1843; https://doi.org/10.1016/j.arthro.2015.03.014.
10 Jeschke GM, et al. Umbilical Cord Lining Membrane and Wharton’s Jelly-Derived Mesenchymal Stem Cells: The Similarities and Differences. Open Tiss. Eng. Regen. Med. J. 2011. 4(1). http://dx.doi.org/10.2174/1875043501104010021.
11 Subramanian A, et al. Comparative Characterization of Cells from the Various Compartments of the Human Umbilical Cord Shows That the Wharton’s Jelly Compartment Provides the Best Source of Clinically Utilizable Mesenchymal Stem Cells. PloS One 10(6) 2015: e0127992; https://dx.doi.org/10.1371%2Fjournal.pone.0127992.
12 Kita K, et al. Isolation and Characterization of Mesenchymal Stem Cells from the Sub-Amniotic Human Umbilical Cord Lining Membrane. Stem Cells Develop. 19(4) 2010: 491–502; https://doi.org/10.1089/scd.2009.0192.
13 Seshareddy K, Troyer D, Weiss ML. Method to Isolate Mesenchymal-Like Cells from Wharton’s Jelly of Umbilical Cord. Methods Cell Biol. 86, 2008: 101–119; https://doi.org/10.1016/s0091-679x(08)00006-x.
14 Choudhery SM, et al. Utility of Cryopreserved Umbilical Cord Tissue for Regenerative Medicine. Curr. Stem Cell Res. Ther. 8(5) 2013: 370–380; https://doi.org/10.2174/1574888×11308050004.
15 Boey PYK. et al. Comparative Study of the Methods of Extracting Mesenchymal Stem Cells from Cryopreserved Wharton’s Jelly. J. Stem Cells Regen. Med. 13(1) 2017: 29; https://dx.doi.org/10.46582%2Fjsrm.1301005.
16 Ryan JM, et al. Mesenchymal Stem Cells Avoid Allogeneic Rejection. J. Inflammation 2(1) 2005: 8; https://doi.org/10.1186/1476-9255-2-8.
17 Le Blanc K. Immunomodulatory Effects of Fetal and Adult Mesenchymal Stem Cells. Cytother. 5(6) 2003: 485–489; https://doi.org/10.1080/14653240310003611.
18 Galipeau J, Sensébé L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell. 22(6) 2018: 824–833; https://doi.org/10.1016/j.stem.2018.05.004.
19 Regmi S, et al. Mesenchymal Stem Cell Therapy for the Treatment of Inflammatory Diseases: Challenges, Opportunities, and Future Perspectives. Eur. J. Cell Biol. 98(5–8) 2019: 151041; https://doi.org/10.1016/j.ejcb.2019.04.002.
20 Minonzio GM, Linetsky E. The Use of Fetal Bovine Serum in Cellular Products for Clinical Applications: Commentary. Cell R46, 2014: 1307; https://www.cellr4.org/wp-content/uploads/sites/2/2014/12/CellR4-2014-2-6-e1307.pdf.
21 NCT04104451: Phase 1, Open-Label Safety Study of Umbilical Cord Lining Mesenchymal Stem Cells (Corlicyte®) to Heal Chronic Diabetic Foot Ulcers. ClinicalTrials.gov, 2020; https://ClinicalTrials.gov/show/NCT04104451.
22 Phan TT. Method of Isolating Mesenchymal Stem Cells from the Amniotic Membrane of the Umbilical Cord: A Mesenchymal Stem Cell Population Isolated from the Amniotic Membrane of the Umbilical Cord and a Cell Culture Medium for Isolating Mesenchymal Stem Cells from the Amniotic Membrane of the Umbilical Cord. USPTO.report 10 May 2018; https://uspto.report/patent/app/20180127721.
23 Chen G, et al. Comparison of Biological Characteristics of Mesenchymal Stem Cells Derived from Maternal-Origin Placenta and Wharton’s Jelly. Stem Cell Res. Ther. 6(1) 2015: 228; https://doi.org/10.1186%2Fs13287-015-0219-6.
24 Gu Y, et al. Changes in Mesenchymal Stem Cells Following Long-Term Culture In Vitro. Molec. Med. Reports 13(6) 2016: 5207–5215; https://doi.org/10.3892/mmr.2016.5169.
25 Moraes DA, et al. A Reduction in CD90 (THY-1) Expression Results in Increased Differentiation of Mesenchymal Stromal Cells. Stem Cell Res. Ther. 7(1) 2016: 97; https://doi.org/10.1186/s13287-016-0359-3.
26 Ode A, et al. CD73/5′-Ecto-Nucleotidase Acts As a Regulatory Factor in Osteo-/Chondrogenic Differentiation of Mechanically Stimulated Mesenchymal Stromal Cells. Eur. Cell Mater. 25(3) 2013; https://doi.org/10.22203/ecm.v025a03.
27 Dominici M, et al. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells: The International Society for Cellular Therapy Position Statement. Cytother. 8(4) 2006: 315–317; https://doi.org/10.1080/14653240600855905.
28 Sargent A, Miller RH. MSC Therapeutics in Chronic Inflammation. Curr. Stem Cell Reports 2(2) 2016: 168–173; https://doi.org/10.1007/s40778-016-0044-6.
29 Wang L-T, et al. Human Mesenchymal Stem Cells (MSCs) for Treatment Towards Immune- and Inflammation-Mediated Diseases: Review of Current Clinical Trials. J. Biomed Sci. 23(1) 2016: 76; https://doi.org/10.1186/s12929-016-0289-5.
30 Maxson S, et al. Concise Review: Role of Mesenchymal Stem Cells in Wound Repair. Stem Cells Translat. Med. 1(2) 2012: 142–149; https://doi.org/10.5966/sctm.2011-0018.
31 Chen L, et al. Paracrine Factors of Mesenchymal Stem Cells Recruit Macrophages and Endothelial Lineage Cells and Enhance Wound Healing. PloS One 3(4) 2008: e1886; https://dx.doi.org/10.1371%2Fjournal.pone.0001886.
32 Shen C, et al. Conditioned Medium from Umbilical Cord Mesenchymal Stem Cells Induces Migration and Angiogenesis. Molec. Med. Reports 12(1) 2015: 20–30; https://doi.org/10.3892/mmr.2015.3409.
Corresponding author Kenny P.Y. Boey (65-6238-0808, [email protected]), Kin Fai Tang, Pengcheng Zhu, Ian Loke, and corresponding author Chi Kwan Leung (65-6238-0808, [email protected]) all are in the laboratory operation group at Cordlife Group Limited, A’Posh Bizhub #06-01/09, 1 Yishun Industrial Street 1, Singapore 768160. Chee Tian Ong, Jeyakumar Masilamani, Ivor J. Lim, and corresponding author Toan Thang Phan (65-6444-9968, [email protected]) all are with CellResearch Corporation Pte. Ltd., 137 Market Street, Unit 08-02, Grace Global Raffles Building, Singapore 048943. KB, CT, JM, and PT conceived and designed the study. KB and JM performed the experiments. KB and CT collected the data. KB, PC, CT and CKL analyzed the data and drafted the manuscript. All authors provided technical guidance, interpreted data, and have read and approved the final manuscript.
You May Also Like