Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.

https://www.istockphoto.com
In BPI’s October 2021 issue, part one of this review introduced the concepts of machine learning (ML) and artificial intelligence (AI), identifying some broad areas of application within vaccine discovery, preclinical testing, and clinical studies. This month, we conclude with a detailed discussion of specific disease targets and AI’s potential in addressing them. Having highlighted the example of Zika virus in part one, below we focus on malaria, tuberculosis, human immunodeficiency virus (HIV), herpesvirus, hepatitis, and pandemic coronavirus.
Malaria
Malaria is a life-threatening infectious disease caused by several species of the protozoa Plasmodium (P. falciparum, P. vivax, P. malariae, and P. ovale) that typically are transmitted by bites of female Anopheles mosquitoes. In 2019, the registered number of malaria cases reached 229 million worldwide, with 94% accounted for in Africa. The death rate has slowly declined in recent decades but remains devastating: more than 400,000 people died in 2019, 67% of them children under five years old. Despite centralized multinational efforts and collaborations to end the global health threat of malaria, the funding gap between actual and needed investment (US$2.6 billion in 2019) continues to increase (65).
Decades of research toward a suitable vaccine resulted in licensure of Mosquirix (RTS,S/AS01) recombinant, preerythrocytic-stage vaccine from GlaxoSmithKline Biologicals SA (GSK). However, the product shows only limited efficacy of 39–50% in children and young infants, however, followed by rapidly waning vaccine-induced immunity (66). Myriad hurdles underlie the failure thus far to generate an efficient vaccine, primarily stemming from the unique biology of the pathogen.
Plasmodium occupies two hosts (mosquitoes and humans) during its life cycle. Three developmental stages occur only in humans: the preerythrocytic (asymptomatic), blood-borne (symptomatic), and gametocytic (transmissible via mosquito bites) stages. Each of those is associated with a distinct antigenic profile and consequently with unique immune-response specificities, accompanied by sophisticated pathogen escape mechanisms (67, 68). In light of the complexities of the malarial pathogen, translation of classical vaccine design methods largely have failed.
With the demand for innovative approaches, it has been hypothesized that the antigen diversity across the multiple malarial strains could be addressed by identification of evolutionally conserved, crossreactive epitopes (69). Such an ambitious approach requires the entire span of computational, structural biology and immunoinformatic tools to align the match of the selected antigen(s) for eliciting specific B- and T-cell responses (70). Published reports have anticipated that immunomics — a combination of immunology, genomics, proteomics, and computational biology — is a promising avenue to generate powerful insights that could accelerate malarial vaccine design. The power of immunomics is to focus on individual characteristics of the parasite in the context of its interaction with a host’s immune system and thus offer a high-throughput, in silico screening method for key target antigens and epitopes (71–73). Advanced technologies also are increasingly leveraged across the malaria vaccine pipeline:
• AI prediction of vaccine outcomes using gene-expression data from clinical-trial subjects (74) and prediction of clinical outcomes (75, 76)
• ML used to identify novel Plasmodium antigens (77), predict vaccine immunogenicity (78, 79), and find opportunities for diagnostic improvement (80, 81).
Table 1 summarizes the current window for widening the applications of AI tools in the context of malarial vaccine improvement and development.
Tuberculosis
Tuberculosis (TB) is a communicable disease that is caused by Mycobacterium tuberculosis and primarily affects the lungs. Despite being potentially a preventable and curable infection, tuberculosis is one of the biggest global health challenges, causing 1.5 million deaths every year. The World Health Organization (WHO) “End Tuberculosis” strategy reduced its incidence only by 9% in 2020 rather than the planned 20% (82). Multiple reasons underlie the obstacles to TB management. They stem from an ineffective range of available diagnostics and inadequate knowledge of reliable latent-stage disease biomarkers, together limiting control of the disease spread (83, 84).
The most desirable way to cope with TB would be an efficient universal vaccine to complement the only licensed pediatric vaccine — against bacillus Calmette–Guérin (BCG) — which was developed 100 years ago and works only for a subset of the population. However, preventing infection, disease, and recurrence would be highly relevant to protect an adolescent population (85, 86). Many efforts now are invested into the development of better TB vaccines, and many vaccine candidates are in clinical trials (85, 87). Nevertheless, the quality and quantity of protective immune responses remain in question. Novel breakthrough technology is needed to provide better diagnostics and ensure prevention and treatment of TB.
Recognizing the unmet need for better TB vaccines, the European Commission funded a project called In Silico Trial for Tuberculosis Vaccine Development (STriTuVaD). Current multidisciplinary consortia want to deliver an in silico platform for simulating TB infections in humans. That would facilitate timely and economical identification of therapeutic and vaccine failures and predict the corresponding solution (88).
One promising approach is offered in the frame of the STriTuVaD project: an agent-based modeling platform called the Universal Immune System Simulator (UISS). It contains all the complexity of biological and molecular entities to resemble closely the biological reality of an immune system while enabling its mathematical description. The current model was extended to mimic a TB infection process, essentially showing different outcomes of the disease depending on the virulence of TB strains and their interaction with a host’s immune system (15, 89).
The mycobacterium pathogen is well recognized for its high level of antibiotic-resistance evolution, which poses a serious threat to global health (82). Multiple groups are attempting to tackle this problem using AI techniques, although most remain in the developmental stage. Thus, AI and ML technologies should be implemented
• to predict antibody-resistant mutations (90, 91)
• to identify highly conserved genome sequences in the frame of antibody-resistant evolution while revealing new drug-resistant mutations through a reference strain-agnostic computational platform (92)
• to define an appropriate drug and mode of administration to treat resistant TB (93, 94).
In the complex fight against TB, early diagnosis and treatment are the basis of restraining disease spread and rescuing lives. We must get past the major hurdle to detecting latent, antibiotic-resistant, nonlung, and pediatric forms of TB. Urgently needed diagnostic approaches require a systematic technological upgrade to move beyond current old-fashioned and often time-consuming techniques with limited accuracy: e.g., the tuberculin skin test (TST), sputum-based test, bacteriological tests, and so on (95–98). With most TB burden restricted to underdeveloped countries, patient access to advanced medical and diagnostic techniques remains limited.
The advantages of digital progress expansion in developed countries should be capitalized upon: e.g., mobile health apps for offering educational materials, preliminary diagnosis, and epidemic surveillance (99). To address insufficiencies in available medical staff for underdeveloped countries, diagnostic confirmation facilitated by AI and ML algorithms specific to TB would be highly desirable, along with increased capacity to process radiological images (87, 100).
Seasonal Influenza
The impacts of seasonal influenza epidemics should not be underestimated. According to WHO, they cause three to five million cases of severe illness every year, with 290,000–650,000 deaths resulting from this respiratory disease (101).
The family of influenza viruses constitutes four virus strains, two of which are responsible for seasonal outbreaks within the human population. Continual antigenic drift of two surface glycoproteins on these viruses leads to evolution of new influenza strains and substrains. So WHO established the Global Influenza Surveillance and Response System (GISRS) to monitor the emergence of drifted influenza virus strains containing surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) with novel antigenic properties. Such monitoring instructs biomanufacturers on which vaccine strain composition to select for each upcoming season. Unfortunately, the approach has only limited success in preventing influenza, with an average vaccine efficacy rate of 19–53% (101–104).
It has been estimated that replacing just 10% of the current annual US vaccination with a “universal flu vaccine” with an efficacy rate of 75% could save $1.1 billion of direct healthcare costs each year (105). A priority of the US National Institutes of Health (NIH) to design a universal influenza vaccine has resulted in 27 candidates tested in clinical trials over the past decade, only three of which made it to phase 3 (106).
Advanced AI technologies can accelerate influenza vaccine development, although such technologies remain in their early development phases. In 2017, Sanofi began a collaboration initiative with Berg (an AI-powered biotechnology company) to identify novel biomarkers of influenza vaccine-driven protective immunity by exploiting AI to analyze patient data from influenza-vaccine clinical trials (107). And one AI breakthrough in influenza vaccine development already has been made by Nikolai Petrovsky (Flinders University professor and research director of Vaxine in Australia). He led the development of the world’s first AI-designed influenza vaccine using a program called Search Algorithm for Ligands (SAM). Results of the phase 1 study are not yet available, however (108, 109).
An earlier attempt by that Australian group applied an in silico structural modeling approach to facilitate the rapid design of a vaccine to mitigate the “swine flu” pandemic in 2009. That record-breaking development led to a successful clinical trial in humans (adjuvanted recombinant Panblok-H1/Advax). Computational modeling provided a deep understanding of the virus structure of that pandemic influenza virus strain as well as the design of Vaxine’s novel Advax adjuvant, which increases vaccine immunogenicity significantly (110, 111). This success demonstrated that exploiting immunoinformatics enables identification of conserved T-cell epitopes — expressed among different seasonal, pandemic, avian and swine influenza virus strains — that can initiate a protective immune response in a transgenic mouse model (112). That provided the starting point for engineering a universal influenza vaccine. Pappalardo, et al. developed a computational pipeline to identify an optimal adjuvant that improves immune response against the influenza virus. In silico design based on simulations of such a response was successfully validated in vivo (113).
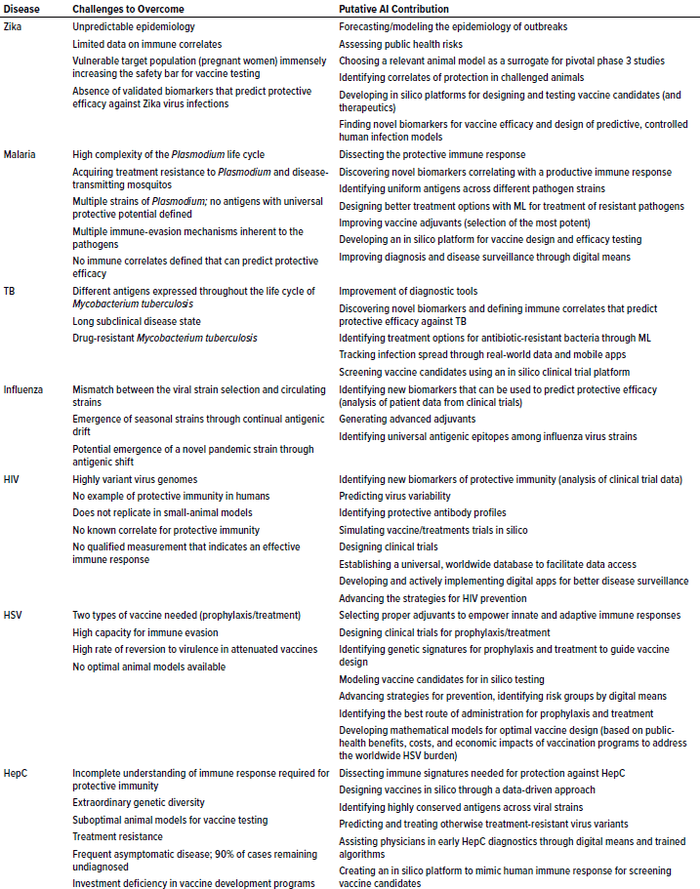
Table 1: Potential AI solutions to tackle problematic, vaccine-preventable diseases of global concern; TB = tuberculosis, HIV = human immunodeficiency virus, HSV = herpes simplex virus, HepC = hepatitis C
Human Immunodeficiency Virus
In the 21st century, HIV continues to be a major global health threat with no available cure or vaccine despite over 30 years of fundamental and clinical research. About 38 million individuals currently live with HIV infections. With significant improvements in access to efficient treatment and prophylactic measures, the rate of newly diagnosed infections and HIV-related deaths has decreased significantly over the past two decades (114). However, the HIV epidemic continues to grow, if at a slowed pace, while the development of a preventive vaccine remains the long-awaited and recommended solution by the International AIDS Society (115, 116).
Development of an effective HIV vaccine has been recognized as an exceptional challenge for decades, so it is not surprising that related fundraising is tenuous (117, 118). Nevertheless, continuing worldwide efforts to develop such a vaccine have identified only a few front-line candidates currently being investigated by efficacy studies in South Africa, with first results expected by the end of 2021 or 2022 (119–121). In-depth discussion of the history of HIV clinical trials is beyond the scope of this review and can be found elsewhere (39, 117). Instead, we highlight attempts to advance the search for a vaccine using AI tools and emphasize areas for possible extension of such methods (Table 1).
Multiple reasons explain the unsuccessful attempts to generate a preventive HIV vaccine so far. The virus exhibits unparalleled genomic variability and immune evasion, thus defying all hopes to benefit from previously successful vaccine technologies. Highly agile viral envelope proteins led researchers to discover the extreme obstacles to inducing broadly neutralizing antibodies. That led to a shift in vaccine developmental programs toward vaccine constructs that mount a potent cellular response against HIV instead (122). Unfortunately, the results of that approach appear to have failed, as exemplified by an adenovirus-based candidate vaccine developed by Merck that initially was considered safe and immunogenic. But a phase 1–2 study demonstrated that individuals with baseline antibody titers specific to adenoviruses who received the candidate vaccine presented a significantly higher HIV burden than did people in the placebo cohort (123).
Despite multiple research advances, the exact mechanism of inducing a functional immune response against HIV infection largely remains a “black box.” It is becoming increasingly clear that no immune components involved in establishing protective immunity should be overlooked. Thus, the whole complexity of the viral interaction with a human immune system must be understood fully, requiring an interdisciplinary approach. That goal is foundational to the European HIV Vaccine Alliance (EHVA), which was initiated in 2016 as a multidisciplinary vaccine platform (MVP) for development and evaluation of HIV vaccines (124). Despite the unmet medical need, however, it seems that this field is integrating innovative technologies at a much lower extent than is cancer drug discovery, for example.
The pipeline of HIV vaccine development has benefited significantly from certain common systems-biology tools:
• mosaic vaccine design (125)
• attempts to dissect a protective immune correlate and identify a specific genetic signature upon vaccination across nonhuman primates, clinical trial participants, and HIV “controllers” (126, 127) — the very small proportion (<1%) of people who live with HIV, do not receive antiretroviral therapy, but spontaneously keep their plasma HIV-1-RNA load below a quantifiable concentration
• identifying vaccine biomarkers (128).
Scientists at Duke University have developed a valuable asset for the vaccinology field (129). They presented a computational tool called ARMADiLLO (antigen receptor mutation analyzer for detection of low-likelihood occurrences), which enabled the evaluation of an improbable B-cell antigen receptor rearrangement required for generation of broadly neutralizing antibodies. Data derived from that tool were functionally validated in an HIV neutralization assay. This platform could provide a powerful means to inform future mutation-guided vaccine design (129).
Specific technologies pushing forward the development of HIV vaccines include systems serology, a high-throughput assay method that enables biophysical and functional profiling of antibodies. Using that platform, Chung et al. performed a comprehensive analysis of neutralizing-antibody profiles among participants of HIV clinical trials. That paves the way toward understanding and dissecting the complexity of an immune response in much more depth than usually can be investigated in clinical trials. That could guide further vaccine development and shed light on an otherwise elusive immune defense mechanism (130).
Application of systems serology provides valuable insights. It has been used in the interpretation of preclinical and clinical data, which could guide improvements through improved understanding vaccine-candidate failures (131–133). Although development of HIV treatments is a tempting route of inquiry, prevention and prophylaxis are most worth applying the magnitude of AI (134).
All those approaches remain in a developmental stage, with significant space for improvement. Table 1 overviews the most promising approaches.
Herpes Simplex
The herpes simplex virus (HSV) is a double-stranded DNA virus that primarily infects humans and is widely distributed across the globe. As of 2016, over 66% of the world population had been infected with HSV1, and over 13% have HSV2 (135). A host becomes a life-long “reservoir” with or without recurrent episodes of viral activation that cause “cold sores” or painful genital ulcer lesions. Both viruses can cause serious neurological complications, particularly through horizontal transmission of HSV2 from mother to child during natural delivery (characterized by a high mortality rate). HSV2 complications in newborns are nearly eliminated in developed countries, however (136). HSV1 in turn is a leading cause of infection-driven blindness (137).
Individuals infected with HSV are often unaware due to the latency phase of the viral life cycle when the virus asymptotically persists in neural ganglia and still is transmittable. Until now, no cure for the infection is available, only symptomatic treatment that limits a person’s viral shedding capacity. In addition to potential quality-of-life reductions, psychosocial implications, and rare but severe complications among infected individuals, note that HSV2-infected women have a three- to sixfold higher risk of acquiring HIV (138). Despite the need for efficient HSV vaccines, a public health priority acknowledged by WHO, they remain unavailable after decades of research (139).
High rates of failure in the late-stage clinical development have made clear that the design of HSV vaccines requires novel approaches and strategies building on the integration of acquired knowledge and advanced technologies (140, 141). WHO has highlighted mathematical modeling implementation as a means to address key gaps in HSV vaccine-design strategy (142). Moreover, two types of vaccines are needed: to prevent initial infection (as a prophylaxis) and to modulate patient immune systems to combat established disease (a therapeutic vaccine). The immense complexity of the combined task opens an infinite avenue for AI to guide balanced and efficient vaccine development (Table 1) (143).
Hepatitis C
HepC is a blood-borne virus distributed around the globe, with over 71 million people who are chronically infected. Severe, life-threatening consequences of chronic hepatitis C infections commonly manifest in hepatocellular carcinoma and liver cirrhosis. Before severe liver pathology is detected, the clinical course of infection is largely asymptomatic, which represents a public-health threat with millions of patients undiagnosed.
In 2016, WHO launched its Global Health Sector Strategy (GHSS) on viral hepatitis, an ambitious program to end the hepatitis epidemic by 2030 (144, 145). The advent of direct-acting antivirals (DAAs) has revolutionized treatment, but drug-resistant virus forms frequently follow such treatment (146, 147). Therefore, a suitable vaccine remains the most desired and preferred option to combat hepatitis C globally with any chance of eliminating the disease. The virus is extraordinarily complicated genetically, with seven to 11 strains, 80 subtypes within those strains, and further viral variants (quasispecies) generated in every infected individual. That maze of genetic variability even outcompetes HIV (148, 149).
Thus, the complicated road toward a hepatitis C vaccine requires transformative innovations from the start: understanding the precise mechanism of a productive immune response and generating suitable small-animal models to test candidate vaccines (150, 151). Although no examples of sterilizing immunity have been recorded among humans, 20–25% of infected people demonstrate spontaneous disease elimination and remain protected from the chronic disease when reexposed and infected (152, 153). Such findings demonstrate that development of a hepatitis C vaccine is feasible — at least one that will protect the population from persistent chronic infections if transmission itself cannot be prevented.
Reported computing technologies mainly focus on the treatment and diagnosis of hepatitis rather than vaccine development. AI and ML technologies have been applied in the context of hepatitis C as follows:
• to predict specific B- and T-cell epitopes using ML algorithms (154–156)
• to design an epitope vaccine using in silico immunoinformatics to engage innate, adaptive, and humoral immunity (157, 158)
• to analyze electronic health records and identify predictive parameters for detecting hepatitis C among patients through ML (159)
• to improve diagnostic modalities with the help of ML (160–163)
• to identify genetic markers associated with responses to DAA therapy (164), genetic viral markers that predict successful DAA therapy (165), and chronic infection outcomes using various ML algorithms (166).
Although the potential economic benefit of a hepatitis C prophylactic vaccine is clear, the funding gap needed to push progress forward limits the speed of R&D progress (167). Therefore, priority must be given to implementation of front-line technologies that can resolve obstacles in time- and cost-efficient vaccine development (see Table 1).
SARS-CoV-2
Following an outbreak in Wuhan, China, at the end of 2019, the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) sped around the world. The battle against the pandemic continues: As of 23 February 2021, the cumulative number of registered COVID-19 cases worldwide reached 110.7 million, with over 2.4 million COVID-19 related deaths (168).
The time has come to prove the utility and accelerate the development of advanced AI technologies in coping with this global crisis. However, in a fast-paced pandemic environment, the major breakthrough in vaccine development was achieved through adaptation of well-established pipelines to meet the demand, although previously they had not succeeded in their intended applications (169). The crisis pushed the border of the imaginable. In particular, it enabled the development and licensure within a year of world-first mRNA based COVID‑19 vaccines from Moderna and from BioNTech and Pfizer — demonstrating 95.4% and 95.6% efficacy, respectively (170–173). The perfect match of the mRNA platform with the biological entity of the virus awakened hopes to end the pandemic, simultaneously raising many issues related to supplying vaccines to the world.
The impact of digitalization indirectly accelerated the success of vaccine development: The SARS-CoV-2 genomic sequence was made widely available immediately after the genome had been sequenced in China. Global efforts to ensure real-time availability and accessibility enabled developers to exploit data-driven approaches immediately and then integrate those insights into therapeutic designs (174–176). One DNA vaccine candidate was designed and optimized within three hours by an advanced computational platform but did not demonstrate significant advantages in studies that followed (177), which led some people to question the superiority of that approach. We speculate that the pandemic has widened the obvious gap between development of AI tools and their actual implementation into vaccine development programs. That raises the necessity of integrating AI tools into early stage product design, which is not feasible in a pandemic situation.
Discussion and Future Perspectives
AI is a promising tool that should accelerate vaccine development and significantly reduce associated costs. In line with scientific progress, the understanding of biological processes becomes increasingly complex. That generates enormous amounts of data to be taken into account in discovery of new vaccines and therapeutics, far exceeding the capacity of the human brain to manage. AI offers great potential to strengthen all vaccine developmental phases (Figure 3 in Part 1), and its impact is not limited to data-mining applications.
Note that AI-driven data-mining, modeling, and prediction tools are beneficial not only for scientists, but also for sponsor companies, public-health agencies, and the epidemiology of pathogens and infectious diseases as a whole. All parties must understand that modern vaccine development no longer belongs to one discipline (vaccinology). It requires close interactions across multiple disciplines — immunology, structural and cellular biology, genetics, and computational engineering — to combine their knowledge and support decision making. The history of vaccine development and licensure suggests that the number of successful market authorizations (MAs) could be increased significantly by empowering science and speeding the discovery of new and effective vaccine antigens. Harnessing AI techniques is one way to bring that process to a new level.
AI tools certainly could shorten the time spent in vaccine development, significantly reducing necessary investments and improving the business sustainability and agility of vaccine-development pipelines. All those acknowledged benefits are just beginning to be realized. The “delay” so far has multiple causes, including the immaturity of the AI toolbox when it comes to modeling of highly sophisticated vaccine-induced immunological processes and interactions (14). Data are the fundamental basis for training and applying computational algorithms. The famous informatics rule “garbage in, garbage out” implies that high-quality, standardized data are necessary to generate meaningful insights. The progress of AI application in vaccine discovery profoundly relies on establishment of accessible, uniform databases — which will require a fundamental global standardization agreement.
Another key problem is represented by an interdisciplinary gap opening during the planning and execution of biological studies. Trained mathematicians and statisticians often become involved during the wrong step in vaccine (product) development, which can result in suboptimal experimental designs and failure to record necessary data appropriately. The quantity of data required should not be underestimated. An advanced deep-learning technology can extract meaningful data patterns automatically only when enough (sometimes millions of) data points are provided for model training (1).
The EU Health Programme 2014–2020 recognizes the difficulty of health-data standardization, handling, and accessibility (178). The overall technological progress seems to be overwhelming, yet the results are integrated only slowly into the thinking of regulatory agencies, medical staff, and even the scientific community. Before new techniques can be implemented, major educational activities are necessary. The European Commission (EC) is training physicians to advance digital literacy, and similar actions should be taken to establish a dialogue between statisticians and other relevant parties (regulatory agencies, stakeholders, and academic institutions) (179). The message and potential of AI applications must get across in an understandable and well-perceived way to ensure optimization of their use (180).
The vaccinology field harbors many issues to be addressed (Table 1). Its development lags behind other life sciences despite the profound medical need. The ongoing pandemic has revealed the problems of vaccine-pipeline modernization to the world. It is imprinted in an EC pharmaceutical strategy report “that innovative approaches to the development, approval and post-authorization monitoring of vaccines and repurposing of medicines are needed” (179). Future perspectives and progress depend on establishment of productive, interdisciplinary consortia — such as the Human Immunomics Initiative — that will apply AI to acceleration of vaccine development (181).
We still have a long way to go in achieving the full potential of AI techniques in vaccine discovery and healthcare as a whole. Nevertheless, a solid base on which to build this breakthrough has been established around the world already.
References
For references 1–64, see Part 1 in BPI’s October 2021 issue.
65 CC BY-NC-SA 3.0 IGO. World Malaria Report 2020: 20 Years of Global Progress and Challenges. World Health Organization: Geneva, Switzerland, 2020.
66 Efficacy and Safety of RTS, S/AS01 Malaria Vaccine With or Without a Booster Dose in Infants and Children in Africa: Final Results of a Phase 3, Individually Randomised, Controlled Trial. Lancet 386(9988) 2015: 31–45.
67 Wilson K, et al. Malaria Vaccines in the Eradication Era: Current Status and Future Perspectives. Exp. Rev. Vaccines 18(2) 2019: 133–151.
68 Lozano JM, et al. The Search of a Malaria Vaccine: The Time for Modified Immuno-Potentiating Probes. Vaccines 9(2) 2021: 115.
69 Mitran CJ, Yanow SK. The Case for Exploiting Cross-Species Epitopes in Malaria Vaccine Design. Front. Immunol. 11, 2020: 335.
70 Ahmadi S, et al. The Role of Digital Technologies in Tackling the Zika Outbreak: A Scoping Review. J. Pub. Health Emerg. 2(6) 2018: 20.
71 De Sousa KP, Doolan DL. Immunomics: A 21st Century Approach to Vaccine Development for Complex Pathogens. Parasitol. 143(2) 2016: 236–244.
72 Doolan DL. Plasmodium Immunomics. Int. J. Parasitol. 41(1) 2011: 3–20.
73 Khan N, et al. An Immunoinformatics Approach to Promiscuous Peptide Design for the Plasmodium falciparum Erythrocyte Membrane Protein-1. Mol. BioSys. 13(10) 2017: 2160–2167.
74 Shayaan A, Ilanchezian I, Rao S. Prediction of Malaria Vaccination Outcomes from Gene Expression Data. Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies: Volume 3 — Bioinformatics. SciTePress: Setúbal, Portugal, 2019: 155–162.
75 Valletta JJ, Recker M. Identification of Immune Signatures Predictive of Clinical Protection from Malaria. PLoS Comput. Biol. 13(10) 2017: e1005812.
76 Proietti C, et al. Immune Signature Against Plasmodium falciparum Antigens Predicts Clinical Immunity in Distinct Malaria Endemic Communities. Mol. Cell. Proteom. 19(1) 2020: 101–113.
77 Liu T, Tang H. A Brief Survey of Machine Learning Methods in Identification of Mitochondria Proteins in Malaria Parasite. Curr. Pharmaceut. Design 26(26) 2020: 3049–3058.
78 Lee EK, et al. Machine Learning for Predicting Vaccine Immunogenicity. Interfaces 46(5) 2016: 368–390.
79 Kazmin D, et al. Systems Analysis of Protective Immune Responses to RTS,S Malaria Vaccination in Humans. Proc. Nat. Acad. Sci. 114(9) 2017: 2425–2430.
80 Poostchi M, et al. Image Analysis and Machine Learning for Detecting Malaria. Translat. Res. 194, 2018: 36–55.
81 Okagbue HI, et al. Diagnosing Malaria from Some Symptoms: A Machine Learning Approach and Public Health Implications. Health Technol. 11(1) 2021: 23–37.
82 CC BY-NC-SA 3.0 IGO. Global Tuberculosis Report 2020. World Health Organization: Geneva, Switzerland, 2020.
83 Walzl G, et al. Tuberculosis: Advances and Challenges in Development of New Diagnostics and Biomarkers. Lancet Infect. Dis. 18(7) 2018: e199–e210.
84 Raviglione M, et al. Scaling Up Interventions to Achieve Global Tuberculosis Control: Progress and New Developments. Lancet 379(9829) 2012: 1902–1913.
85 McShane H. Insights and Challenges in Tuberculosis Vaccine Development. Lancet Resp. Med. 7(9) 2019: 810–819.
86 McNerney R, et al. Tuberculosis Diagnostics and Biomarkers: Needs, Challenges, Recent Advances, and Opportunities. J. Infect. Dis. 205(sup 2) 2012: S147–S58.
87 Meraj SS, et al. Artificial Intelligence in Diagnosing Tuberculosis: A Review. Int. J. Adv. Sci. Eng. IT 9(1) 2019: 81–91.
88 EU-India Cooperation on Health Challenge Posed By Tuberculosis in India. STriTuVaD Consortium: Sheffield, UK 2020.
89 Pappalardo F, et al. An Agent Based Modeling Approach for the Analysis of Tuberculosis–Immune System Dynamics. International Conference on Bioinformatics and Biomedicine. Madrid, Spain, 3–6 December 2018. IEEE: Piscataway, NJ.
90 Jamal S, et al. Artificial Intelligence and Machine Learning Based Prediction of Resistant and Susceptible Mutations in Mycobacterium tuberculosis. Scientif. Rep. 10(1) 2020: 1–16.
91 Kouchaki S, et al. Application of Machine Learning Techniques to Tuberculosis Drug Resistance Analysis. Bioinformatics 35(13) 2019: 2276–2282.
92 Kavvas ES, et al. Machine Learning and Structural Analysis of Mycobacterium tuberculosis Pan-Genome Identifies Genetic Signatures of Antibiotic Resistance. Nature Communic. 9(1) 2018: 1–9.
93 Deshpande D, et al. Levofloxacin Pharmacokinetics/Pharmacodynamics, Dosing, Susceptibility Breakpoints, and Artificial Intelligence in the Treatment of Multidrug-Resistant Tuberculosis. Clin. Infect. Dis. 67(sup 3) 2018: S293–S302.
94 Modongo C, et al. Artificial Intelligence and Amikacin Exposures Predictive of Outcomes in Multidrug-Resistant Tuberculosis Patients. Antimicrob. Agents Chemother. 60(10) 2016: 5928–5932.
95 Pai M, Kalantri S, Dheda K. New Tools and Emerging Technologies for the Diagnosis of Tuberculosis: Part I, Latent Tuberculosis. Exp. Rev. Molec. Diagnost. 6(3) 2006: 413–422.
96 Pai M, Kalantri S, Dheda K. New Tools and Emerging Technologies for the Diagnosis of Tuberculosis: Part II, Active Tuberculosis and Drug Resistance. Exp. Rev. Molec. Diag. 6(3) 2006: 423–432.
97 Drain PK, et al. Guidance for Studies Evaluating the Accuracy of Biomarker-Based Nonsputum Tests to Diagnose Tuberculosis. J. Infect. Dis. 220(sup 3) 2019: S108–S15.
98 Hesseling AC, et al. A Critical Review of Diagnostic Approaches Used in the Diagnosis of Childhood Tuberculosis. Int. J. Tuberc. Lung Dis. 6(12) 2002: 1038–1045.
99 Keutzer L, Wicha SG, Simonsson US. Mobile Health Apps for Improvement of Tuberculosis Treatment: Descriptive Review. JMIR mHealth uHealth 8(4) 2020: e17246.
100 Dande P, Samant P. Acquaintance to Artificial Neural Networks and Use of Artificial Intelligence As a Diagnostic Tool for Tuberculosis: A Review. Tuberculosis 108, 2018: 1–9.
101 Influenza Update No. 331. World Health Organization: Geneva, Switzerland, 24 December 2018.
102 Biswas A, Chakrabarti AK, Dutta S. Current Challenges: From the Path of “Original Antigenic Sin” Towards the Development of Universal Flu Vaccines: Flu Vaccine Efficacy Encounters Significant Hurdles from Pre-Existing Immunity of the Host Suggesting Assessment of Host Immunity Before Vaccination. Int. Rev. Immunol. 39(1) 2020: 21–36.
103 Jackson ML, et al. Influenza Vaccine Effectiveness in the United States During the 2015–2016 Season. New Eng. J. Med. 377(6) 2017: 534–543.
104 McLean HQ, et al. Influenza Vaccine Effectiveness in the United States During 2012–2013: Variable Protection By Age and Virus Type. J. Infect. Dis. 211(10) 2015: 1529–1540.
105 Sah P, et al. Future Epidemiological and Economic Impacts of Universal Influenza Vaccines. Proc. Nat. Acad. Sci. 116(41) 2019: 20786–20792.
106 Corder BN, et al. A Decade in Review: A Systematic Review of Universal Influenza Vaccines in Clinical Trials During the 2010 Decade. Viruses 12(10) 2020: 1186.
107 Li A. Precision Flu Vaccines? Sanofi Inks AI Pact to Find Potential Biomarkers 2017. Fierce Pharma 31 October 2017.
108 Petrovsky N. Vaxine. Hum. Vacc. Immunother. 12(11) 2016: 2726–2728.
109 Masige H. Australian Researchers Have Just Released The World’s First AI-Developed Vaccine. Business Insider 13 July 2019.
110 Honda-Okubo Y, et al. Panblok-H1+ Advax H1N1/2009pdm Vaccine: Insights into Rapid Development of a Delta Inulin Adjuvanted Recombinant Pandemic Influenza Vaccine. Hum. Vacc. Immunother. 13(6) 2017: 1261–1271.
111 Honda-Okubo Y, Saade F, Petrovsky N. Advax™, a Polysaccharide Adjuvant Derived from Delta Inulin, Provides Improved Influenza Vaccine Protection Through Broad-Based Enhancement of Adaptive Immune Responses. Vaccine 30(36) 2012: 5373–5381.
112 Eickhoff CS, et al. Highly Conserved Influenza T Cell Epitopes Induce Broadly Protective Immunity. Vaccine 37(36) 2019: 5371–5381.
113 Pappalardo F, et al. A Computational Model to Predict the Immune System Activation By Citrus-Derived Vaccine Adjuvants. Bioinformatics 32(17) 2016: 2672–2680.
114 HIV/AIDS. World Health Organization: Geneva, Switzerland, 2020.
115 Corey L, Gray GE. Preventing Acquisition of HIV is the Only Path to an AIDS-Free Generation. Proc. Nat. Acad. Sci. 114(15) 2017: 3798–3800.
116 Selinger C, et al. Targeting and Vaccine Durability Are Key for Population-Level Impact and Cost-Effectiveness of a Pox-Protein HIV Vaccine Regimen in South Africa. Vaccine 37(16) 2019: 2258–2267.
117 Jones LD, Moody MA, Thompson AB. Innovations in HIV-1 Vaccine Design. Clin. Therapeut. 42(3) 2020: 499–514.
118 Shapiro SZ. HIV Vaccine Development: 35 Years of Experimenting in the Funding of Biomedical Research. Viruses 12(12) 2020: 1469.
119 Bekker L-G, et al. The Complex Challenges of HIV Vaccine Development Require Renewed and Expanded Global Commitment. Lancet 395(10221) 2020: 384–388.
120 Russell ND, Marovich MA. Pox-Protein Public Private Partnership Program and Upcoming HIV Vaccine Efficacy Trials. Curr. Opin. HIV AIDS 11(6) 2016: 614–619.
121 Pitisuttithum P, Marovich MA. Prophylactic HIV Vaccine: Vaccine Regimens in Clinical Trials and Potential Challenges. Exp. Rev. Vaccines 19(2) 2020: 133–142.
122 Burton DR, et al. HIV Vaccine Design and the Neutralizing Antibody Problem. Nature Immunol. 5(3) 2004: 233–236.
123 Sekaly R-P. The Failed HIV Merck Vaccine Study: A Step Back or a Launching Point for Future Vaccine Development? J. Exper. Med. 205(1) 2008: 7–12.
124 European HIV Vaccine Alliance (EHVA). A EU Platform for the Discovery and Evaluation of Novel Prophylactic and Therapeutic Vaccine Candidates. European Commission: Berlin, Germany, 2016.
125 Barouch DH, et al. Evaluation of a Mosaic HIV-1 Vaccine in a Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 1/2a Clinical Trial (APPROACH) and in Rhesus Monkeys (NHP 13-19). Lancet 392(10143) 2018: 232–243.
126 Ehrenberg PK, et al. A Vaccine-Induced Gene Expression Signature Correlates with Protection Against SIV and HIV in Multiple Trials. Sci. Translat. Med. 11(507) 2019: eaaw4236.
127 Martin-Gayo E, et al. Immunological Fingerprints of Controllers Developing Neutralizing HIV-1 Antibodies. Cell Reports 30(4) 2020: 984–996.
128 Janes HE, et al. Higher T-Cell Responses Induced By DNA/rAd5 HIV-1 Preventive Vaccine Are Associated with Lower HIV-1 Infection Risk in an Efficacy Trial. J. Infect. Dis. 215(9) 2017: 1376–1385.
129 Wiehe K, et al. Functional Relevance of Improbable Antibody Mutations for HIV Broadly Neutralizing Antibody Development. Cell Host Microbe. 23(6) 2018: 759–765.
130 Chung AW, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity Against HIV Using Systems Serology. Cell 163(4) 2015: 988–998.
131 Barouch DH, et al. Protective Efficacy of Adenovirus/Protein Vaccines Against SIV Challenges in Rhesus Monkeys. Science 349(6245) 2015: 320–324.
132 Bradley T, et al. Pentavalent HIV-1 Vaccine Protects Against Simian-Human Immunodeficiency Virus Challenge. Nature Commun. 8(1) 2017: 1–15.
133 Vaccari M, et al. Adjuvant-Dependent Innate and Adaptive Immune Signatures of Risk of SIV mac251 Acquisition. Nature Med. 22(7) 2016: 762–770.
134 Marcus JL, et al. Artificial Intelligence and Machine Learning for HIV Prevention: Emerging Approaches to Ending the Epidemic. Curr. HIV/AIDS Rep. 17(3) 2020: 171–179.
135 James C, et al. Herpes Simplex Virus: Global Infection Prevalence and Incidence Estimates, 2016. Bull. WHO 98(5) 2020: 315.
136 Pinninti SG, Kimberlin DW. Maternal and Neonatal Herpes Simplex Virus Infections. Am. J. Perinatol. 30(02) 2013: 113–120.
137 Koujah L, Suryawanshi RK, Shukla D. Pathological Processes Activated By Herpes Simplex Virus-1 (HSV-1) Infection in the Cornea. Cell. Molec. Life Sci. 76(3) 2019: 405–419.
138 Looker KJ, et al. Global and Regional Estimates of the Contribution of Herpes Simplex Virus Type 2 Infection to HIV Incidence: A Population Attributable Fraction Analysis Using Published Epidemiological Data. Lancet Infect. Dis. 20(2) 2020: 240–249.
139 Gottlieb SL, et al. Meeting Report: Initial World Health Organization Consultation on Herpes Simplex Virus (HSV) Vaccine Preferred Product Characteristics, March 2017. Vaccine 37(50) 2019: 7408–7418.
140 Kaufmann JK, Flechtner JB. Evolution of Rational Vaccine Designs for Genital Herpes Immunotherapy. Curr. Opin. Virol. 17, 2016: 80–86.
141 Kim HC, Lee HK. Vaccines Against Genital Herpes: Where Are We? Vaccines 8(3) 2020: 420.
142 Gottlieb SL, et al. Modelling Efforts Needed to Advance Herpes Simplex Virus (HSV) Vaccine Development: Key Findings from the World Health Organization Consultation on HSV Vaccine Impact Modelling. Vaccine 37(50) 2019: 7336–7345.
143 Spicknall IH, et al. Review of Mathematical Models of HSV-2 Vaccination: Implications for Vaccine Development. Vaccine 37(50) 2019: 7396–7407.
144 Global Health Sector Strategies on Viral Hepatitis 2016-2021: Towards Ending Viral Hepatitis. World Health Organization: Geneva, Switzerland, 2016.
145 Waheed Y, et al. Hepatitis Elimination By 2030: Progress and Challenges. World J. Gastroenterol. 24(44) 2018: 4959.
146 Franco S, et al. Detection of a Sexually Transmitted Hepatitis C Virus Protease Inhibitor-Resistance Variant in a Human Immunodeficiency Virus–Infected Homosexual Man. Gastroenterol. 147(3) 2014: 599–601.
147 Sarrazin C, Zeuzem S. Resistance to Direct Antiviral Agents in Patients with Hepatitis C Virus Infection. Gastroenterol. 138(2) 2010: 447–462.
148 Bailey JR, Barnes E, Cox AL. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology 156(2) 2019: 418–430.
149 Alexopoulou A. Genetic Heterogeneity of Hepatitis C Virus and Its Clinical Significance. Annals Gastroenterol. 14(4) 2001: 261–272.
150 Thomas E, Liang TJ. Experimental Models of Hepatitis B and C: New Insights and Progress. Nature Rev. Gastroenterol. Hepatol. 13(6) 2016: 362–374.
151 Li D, Huang Z, Zhong J. Hepatitis C Virus Vaccine Development: Old Challenges and New Opportunities. Nat. Sci. Rev. 2(3) 2015: 285–295.
152 Sacks-Davis R, et al. Hepatitis C Virus Reinfection and Spontaneous Clearance of Reinfection: The InC3 Study. J. Infect. Dis. 212(9) 2015: 1407–1419.
153 Osburn WO, et al. Spontaneous Control of Primary Hepatitis C Virus Infection and Immunity Against Persistent Reinfection. Gastroenterol. 138(1) 2010: 315–324.
154 Prabdial-Sing N, Puren AJ, Bowyer SM. Sequence-Based In Silico Analysis of Well Studied Hepatitis C Virus Epitopes and Their Variants in Other Genotypes (Particularly Genotype 5a) Against South African Human Leukocyte Antigen Backgrounds. BMC Immunol. 13(1) 2012: 1–15.
155 Huang W-L, et al. Prediction of Linear B-Cell Epitopes of Hepatitis C Virus for Vaccine Development. BMC Med. Genomics 8(4) 2015: 1–13.
156 Joshi A, et al. Application of HMM-Viterbi Model for Identification of Epitopic Signature Within Screened Protein-Antigens of Hepatitis C Virus. Eur. J. Molec. Clin. Med. 7(7) 2020: 4095–4102.
157 Khalid H, Ashfaq UA. Exploring HCV Genome to Construct Multi-Epitope Based Subunit Vaccine to Battle HCV Infection: Immunoinformatics Based Approach. J. Biomed. Informatics 108, 2020: 103498.
158 Dehghan Z, et al. Development of Polyepitopic Immunogenic Contrast Against Hepatitis C Virus 1a-6a Genotype By In Silico Approach. Biomed. Biotechnol. Res. J. 4(4) 2020: 355.
159 Chicco D, Jurman G. An Ensemble Learning Approach for Enhanced Classification of Patients with Hepatitis and Cirrhosis. IEEE Access 9, 2021: 24485–24498.
160 Ahammed K, et al. Predicting Infectious State of Hepatitis C Virus Affected Patients Applying Machine Learning Methods. IEEE Region 10 Symposium. Dhaka, Bangladesh, 5–7 June 2020. IEEE: Piscataway, NJ.
161 Akella AB, Akella S. Applying Machine Learning to Evaluate for Fibrosis in Chronic Hepatitis C. medRxiv 4 November 2020.
162 Orooji A, Kermani F. Machine Learning Based Methods for Handling Imbalanced Data in Hepatitis Diagnosis. Front. Health Informatics 10(1) 2021: 57.
163 Zhang Z-M, et al. Early Diagnosis of Hepatocellular Carcinoma Using Machine Learning Method. Front. Bioeng. Biotechnol. 8, 2020: 254.
164 KayvanJoo AH, Ebrahimi M, Haqshenas G. Prediction of Hepatitis C Virus Interferon/Ribavirin Therapy Outcome Based on Viral Nucleotide Attributes Using Machine Learning Algorithms. BMC Res. Notes 7(1) 2014: 1–11.
165 Haga H, et al. A Machine Learning-Based Treatment Prediction Model Using Whole Genome Variants of Hepatitis C Virus. PloS One 15(11) 2020: e0242028.
166 Ioannou GN, et al. Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients with Hepatitis C Cirrhosis. JAMA Network Open 3(9) 2020: e2015626-e.
167 Scott N, et al. The Case for a Universal Hepatitis C Vaccine to Achieve Hepatitis C Elimination. BMC Med. 17(1) 2019: 1–12.
168 Weekly Epidemiological Update. World Health Organization: Geneva, Switzerland, 23 February 2021.
169 Dolgin E. Unlocking the Potential of Vaccines Built on Messenger RNA. Nature 574(7778) 2019: S10–S12.
170 Polack FP, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New Eng. J. Med. 383(20) 2020: 2603–2615.
171 Rubin EJ, Longo DL. SARS-CoV-2 Vaccination — An Ounce (Actually, Much Less) of Prevention. New Eng. J. Med. 383(20) 2020: 2677–2678.
172 Anderson EJ, et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. New Eng. J. Med. 383(20) 2020: 2427–2438.
173 Jackson LA, et al. An mRNA Vaccine Against SARS-CoV-2: Preliminary Report. New Eng. J. Med. 383(20) 2020: 1920–1931.
174 Defendi HGT, da Silva Madeira L, Borschiver S. Analysis of the COVID-19 Vaccine Development Process: An Exploratory Study of Accelerating Factors and Innovative Environments. J. Pharmaceut. Innov. 2 February 2021: 1–17.
175 Liu C-H, et al. Highlight of Severe Acute Respiratory Syndrome Coronavirus-2 Vaccine Development Against COVID-19 Pandemic. J. Chinese Med. Assoc. 84(1) 2021: 9–13.
176 Waltz E. What AI Can — and Can’t — Do in the Race for a Coronavirus Vaccine. IEEE Spectrum 29 September 2020.
177 Smith TR, et al. Immunogenicity of a DNA Vaccine Candidate for COVID-19. Nature Commun. 11(1) 2020: 1–13.
178 DG Health and Food Safety. Assessment of the EU Member States’ Rules on Health Data in the Light of GDPR. European Union: Luxembourg, 2021.
179 Joint Statement: Training Future-Proof Doctors for the Digital Society. European Union: Luxembourg, December 2020.
180 Schuerman L. RTS, S Malaria Vaccine Could Provide Major Public Health Benefits. Lancet 394(10200) 2019: 735–736.
181 Human Immunomics Initiative Will Decode Immune System, Speed New Vaccines. Harvard T.H. Chan School of Public Health: Boston, MA, 14 April 2020.
Aleksandra Nevmerzhitskaya is a consultant, Mariya Gromova is a senior consultant, and Dr. Michael Pfleiderer is a principal consultant at Biopharma Excellence (a PharmaLex company), Munich Technology Center, Agnes-Pockels-Bogen 1, 80992 Munich, Germany; [email protected].
You May Also Like