Accelerating Process Development Through Flexible Automated Workflows
November 17, 2017
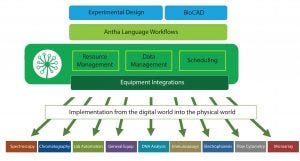
Figure 1: Layers of the Antha software; workflows are detailed in the Antha language then compiled to low-level instructions by the AnthaOS before being pushed through drivers to directly interface with diverse lab equipment.
Synthace began as a bioprocess optimization company in 2011, spun out of University College, London. The company worked on multifactorial approaches with 15–30 factors simultaneously instead of seven or eight. The work investigated genetic strain engineering factors alongside process parameters, defining deep interactions between the way strains were designed and the way they were treated in bioprocesses. Those complex experiments gave unique insight into the complexities of biological processes, but they were exceptionally taxing to plan and carryout manually. Automation seemed to be a solution, but programming the liquid handling steps necessary for this type of experiment was similarly arduous.
Software for Integration and Control
Hardware for laboratory automation is exceptionally good, but control over it can be complex and time consuming. This needn’t be the case. A helpful analogy is that of a smart phone. Smart phones are collections of sophisticated hardware modules — from GPS systems to powerful computer processing units, graphics processing units, touchscreens, and cameras. However, most people tend to think of phones as highly integrated — largely because the software that lies above the hardware presents a highly intuitive interface while it orchestrates the diverse functions of the hardware for a given task.
Antha software was designed as the equivalent software layer for biology. The goal was to make the process of achieving precise control over laboratory equipment as intuitive and to help users perform complicated experiments. The Antha language (AnthaLang) includes the vocabulary required to perform biology experiments, thus allowing it to model the logic of liquids, concentrations, labware, and physical actions. Workflows encoded in the AnthaLang language describe the movement of physical samples and data at a high level and can be compiled down by the Antha operating system to all low-level instructions that are then required to carry out a given experiment (Figure 1). A driver layer then converts those low-level instructions through to specific instructions required by laboratory hardware. My company has demonstrated that this process gives equivalent biological functionality across different makes and manufacturers of laboratory automation.
The idea is to bring together biology and the digital world. Biological issues such as transferability and reproducibility of methodologies, traceability, and dealing with complexity become routine when workflows are defined in code. When a process is digitally defined, technical transfer involves only the transfer of a file. All automated processes programmed in this way are fully logged and traceable. Complex experiments often become as easy to carry out as simple ones.
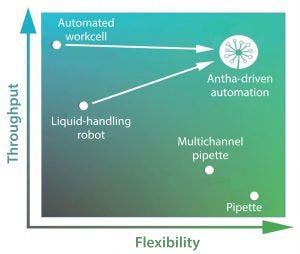
Figure 2: Experiments that properly address biological complexity are tough to perform. For example, liquid-handling automation can be arduous to program, meaning it can be faster to do complex experiments by hand. This is limited in throughput, preventing biologists from properly addressing the complexities of biological systems. The Antha software enables highly flexible programming of laboratory automation, giving high throughput of complex experiments.
Experiment Flexibility and Throughput
Performing experiments and workflows are typically thought of in terms of flexibility and throughput (Figure 2). Carrying out an experiment with a pipette offers ultimate flexibility, but throughput is low. Throughput can increase eightfold with the use of a multichannel pipette, but doing so reduces flexibility somewhat because now a researcher must think about an experimental design that allows the use of eight channels in a row. A liquid-handling robot can provide even higher throughput, but it substantially lowers flexibility because of the necessity to plan and program-in every liquid-handling step. Adaptability of programmed methods is low, with minor changes often requiring large proportions of the method to be reprogrammed. Ultimate throughput comes from fully automated work cells, but they are so fastidious and complex to program that only a few specialized experts work with such systems.
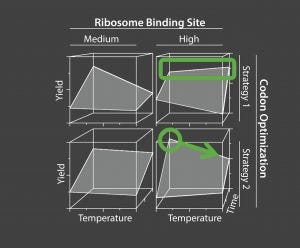
Figure 3: Example of a model from 576 experiments and investigating 27 factors.
The Antha software was designed so that the programming of flexible workflows becomes much easier. When optimizing a biologics production platform in collaboration with Merck, my company used the software to conduct high-dimensional optimizations.
Figure 3 is a model from the third iteration of a multifactorial experimentation, a total of 576 separate experiments. It shows a four-factor interaction between genetic factors (ribosomal binding site strength and codon optimization) and process parameters (time and temperature), which can be identified and defined only through complex high-dimensional experimentation.
Workflow Case Study: Genetic Construct Assembly
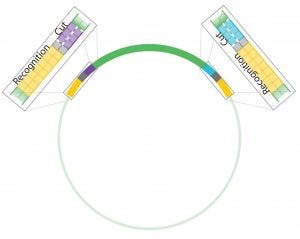
Figure 4: Antha construct assembly uses Type IIS restriction enzymes (T2S), which are known as “remote cutters” because they cut away from their recognition sites. This allows an experimentalist to choose overhang sequences and flank genetic parts of interest with the restriction enzyme recognition sites so that they can be removed after digestion.
One of the workflows that has coded into the Antha software is a highly optimized plasmid assembly method. It is based on a Golden Gate cloning, which uses Type IIS restriction enzymes (Figures 4 and 5). When the method was first trialed in a laboratory, it was complex and had low reliability. However, after multifactorial optimization and characterization of the method using the Antha software, a robust and simplified workflow was engineered. That workflow enabled automation on even simple liquid-handling robots. Running the workflow through the Antha software enables flexible reprogramming of liquid-handling automation irrespective of the details of the genetic designs.
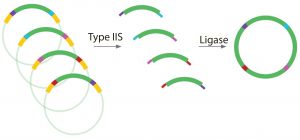
Figure 5: Complementary overhangs and T2S restriction enzyme sites can be designed into DNA fragments, allowing multiple DNA fragments to be ligated together in one “pot.” Example applications include shuffling multiple antibody CDRs and constant regions; fusion protein investigations; leader sequence investigations; promoter sequence and RBS strength investigations; and combinatorial library generations.
For this example, a design file detailing 24 assemblies was uploaded into the Antha software, which then dynamically computed all low-level liquid-handling steps that were required to automatically produce those assemblies in a laboratory (Figure 6). The software handled all low-level planning and then pushed those instructions through a driver to the liquid-handling robot in the laboratory, which carried out the assemblies. The drivers are exceptionally important, because they enable the Antha software to talk to the diversity of laboratory equipment. My company has demonstrated equivalent functionality across different makes and models of robots.
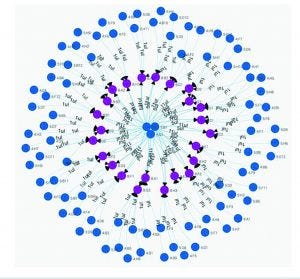
Figure 6: Liquid-handling action of Antha system; blue dots are source wells and purple dots are destination wells.
Because workflows in the Antha software are unambiguously coded and carried out with automation, there is no room for misinterpretation of instructions, assumed knowledge, or operator-to-operator variability as is common when processes need to be transferred. This means that it is possible to get instant, perfect transfer of a method into a different laboratory located even 3,500 miles away. When my company transferred the assembly process to Merck, their biologists gained our sophisticated genetic manipulation capabilities instantly. Antha-driven assembly increased their throughput by an order of magnitude and enabled an estimated 25% reduction in overall vector construction timelines, a 33% reduction in costs, while dramatically reducing manual steps and increasing robustness. That enabled the rapid investigation of large arrays of genetic options alongside process factors for full bioprocess optimization.
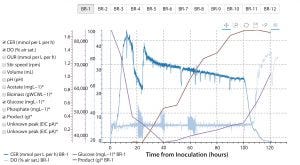
Figure 7: Automatically generated online/offline bioprocess data plot in the Antha software; graph shows interactive interface, which has been used to show the carbon dioxide evolution rate (CER), dissolved oxygen (DO), glucose, and product levels in bioreactor one (BR-1) of a 12-bioreactor ambr250 run. ( * = offline data)
Data Integration and Analysis: Toward Dynamic Process Analytics and Control
Full digital integration (possible with the Antha software) is of particular value for analytics. The software provides reproducibility, and every sample is fully tracked through the system. Data parsed by the Antha software from analytical equipment are automatically associated with the exact samples and sample-processing workflows that come before analysis — thereby automatically generating structured data that can be rapidly analyzed and visualized. With the structured data, analysts have interactive online/offline data plots automatically generated (Figure 7), instead of requiring days or weeks of work to collate, reformat, and display results from bioreactors and diverse analytics with conventional tools.
As the needs of the industry shift to ever increasing sophistication of analytics and control, rapid, cloud-based generation, storage, and analysis of bioprocess data will become essential. For example, Synthace is currently engaged in projects that are looking to apply the Antha software to gene and cell therapies, aiming for full integration of those bioprocesses into the digital world to enable dynamic, high-dimensional process analysis and control.
Markus Gershater is chief scientific officer at Synthace; [email protected].
This article is based on a presentation at Biotech Week Boston, 24–28 September 2017.
You May Also Like





