- Sponsored Content
- Validation
- Laboratory Equipment
- QA/QC
Qualitative and Quantitative Host Cell Protein Analysis Using Mass Spectrometry
Sponsored by BioPharmaSpec
 Host cell proteins (HCPs) originate from host organisms that are used to produce biopharmaceutical products. They are in-process contaminants that must be minimized during downstream process operations. According to regulatory agencies, the maximum permitted level of total HCP in a biopharmaceutical product is 100 ng per mg (100 ppm) (1). HCPs can decrease drug efficacy and pose a risk to patient safety because they can bring on undesirable immune responses. Thus, HCPs are a critical quality attribute that should be minimized and closely monitored according to US (21 CFR 610.13, USP <1132>), European (ICH Q6B, EMA 3AB1A), and other regulatory guidelines (2–5).
Host cell proteins (HCPs) originate from host organisms that are used to produce biopharmaceutical products. They are in-process contaminants that must be minimized during downstream process operations. According to regulatory agencies, the maximum permitted level of total HCP in a biopharmaceutical product is 100 ng per mg (100 ppm) (1). HCPs can decrease drug efficacy and pose a risk to patient safety because they can bring on undesirable immune responses. Thus, HCPs are a critical quality attribute that should be minimized and closely monitored according to US (21 CFR 610.13, USP <1132>), European (ICH Q6B, EMA 3AB1A), and other regulatory guidelines (2–5).
Historically, analysis of HCPs has been performed using enzyme-linked immunosorbent assay (ELISA) tests based on polyclonal antibodies (generated to the whole host cell proteome). This approach enables detection and quantitation of HCPs as a population. Good reproducibility, sensitivity, and high throughput are some advantages of using ELISA methods. However, drawbacks include lack for sensitivity of low-reactive HCPs, long method development times, and an inability to recognize individual HCPs in a drug product without production of HCP-specific monoclonal antibodies. ELISA methods can detect only ~70% of HCPs potentially present in a sample.
Comparatively, an analysis based on proteomic mass spectrometry (MS) can identify de novo a wider population of individual HCPs, without the need for specialized antibody reagents or product- and process-specific method development. MS is an orthogonal technique to ELISA and thus expands the range of HCPs detectable in a sample (3).
A Phased Approach to HCP Analysis
HCP analysis using MS is performed as a phased approach, which allows identification and quantitation of HCPs.
Phase 1, Qualitative Analysis to Identify Protein Impurities Present in Each Sample: Samples of total HCP extract (cell lysate) and of a drug product from multiple steps of a downstream purification process should be analyzed. This analysis can be used to detect potential HCPs present in the drug product and to monitor HCP profile changes during purification process steps.
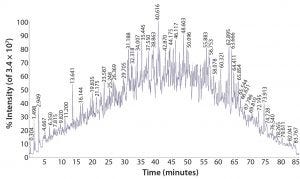
Figure 1: Total ion-current chromatogram from LC-ESI-MS analysis of a proteolytically digested K562 lysate standard
HCP extract is proteolytically digested and analyzed using direct on-line liquid chromatography, electrospray ionization, mass spectrometry (LC/ESI-MS/MS) with a high-resolution, high–mass-accuracy quadrupole time-of-flight (Q-TOF) instrument (Figure 1). This technique generates fragment ions that can be used to determine the identity of peptides and thus identify the proteins from which they are derived. Several replicate analyses are performed to ensure the creation of a robust protein spectral library specific to a given manufacturing process used to produce a biopharmaceutical product. If necessary, two-dimensional LC or gel-based methods can be developed for in-depth peptide and protein library characterization.
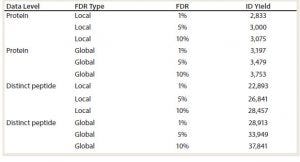
Table 1: Summary of protein and distinct peptide identifications found in a K562 human cell lysate standard as calculated with proteomic software; global level information represents the estimated number of correct identifications at the indicated false discover rate (FDR). Local level information represents the estimated number of high confidence identifications (ID yield) at the indicated FDRs.
LC/ESI-MS/MS data are analyzed against a relevant protein sequence database using bioinformatics proteomics software. Application of stochastic (decoy) sequence databases allows for monitoring false discovery rates (FDR), which are estimated as a number of incorrect protein identifications (from the decoy database) and is used to characterize method performance. In the example shown in Table 1, the number of proteins found at a global 1% FDR — an estimate that 1% of protein identifications (IDs) are false — is 3,197. Using FDR analysis, a spectral library is built based on the most reliable peptide and proteins IDs.
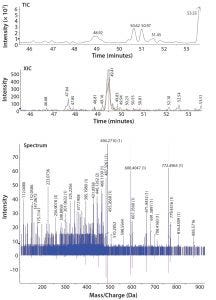
Figure 2: Total ion-current (TIC) chromatogram of a proteolytically digested MAb standard spiked with 20 ppm of myoglobin (top); extracted ion chromatogram (XIC) of a 20-ppm myoglobin spike peptide (middle); and fragmentation spectra fitted to the library spectrum confirming the presence of a specific myoglobin peptide (bottom)
Following assessment of the full HCP library, a drug product at different purification steps will undergo proteolytic digestion and LC/ESI-MS/MS analysis. Data are screened against the product-specific HCP spectral library using proteomic software (Figure 2). That process allows for detection of HCPs to <10 ppm amounts. The spectral library screening technique allows detection of low-level HCPs that might not be detected readily using other methods. Then bioinformatics tools are used to filter the data, followed by manual data inspection to confirm reported HCPs.
Phase 2, Relative Quantitation Against the Response from a Set of Standard Proteins Spiked into a Sample: HCP quantitation is performed following addition of a set of supplementary proteins at known concentrations to estimate the levels of protein impurities in a sample. HCPs present in a biopharmaceutical product are identified and quantified based on LC/ESI-MS/MS data analyzed against the updated HCP protein library and the relevant protein database supplemented with the respective protein sequences. It is important to note that quantitation against supplementary protein standards will provide a quantitation estimate relative to those protein standards. That estimate will be compromised to an extent by the differences in ionization efficiency among different peptides in a sample and reference protein mixtures. More accurate quantitation methods based on MS can be used for all HCPs of particular interest after initial screening.
Phase 3, Quantitation Against Heavy-Labeled Marker Peptides: To provide accurate quantitation of identified protein impurities, including HCPs of specific interest, heavy-labeled peptide analogues of HCP marker peptides identified during phases 1 and 2 can be synthesized and spiked into samples. Amounts of the HCPs of interest are then calculated based on comparing the MS response from the native peptides and the known concentration of heavy-isotope–labeled HCP-specific (marker) peptides.
Multiple reaction monitoring (MRM) methods based on assessment of MS signal filtered by both a peptide parent and fragment ion mass are developed for each native and heavy-isotope–labeled marker peptide on a triple-quadrupole mass spectrometer. These specific MRM transitions can be used to detect and quantitate each peptide of interest for HCPs and their heavy-isotope–labeled standards. Triple-quadrupole mass spectrometers are useful for this analysis because they have a relatively large linear response range and relatively high sensitivity and selectivity.
A Complete Analysis
Regulators are requesting orthogonal approaches to HCP analysis — but not replacements for the HCP-ELISA tests that are still required — to provide an increased breadth of HCP identification. MS provides identification and quantitation of specific HCPs present in drug product samples and is not limited to HCPs that produce immunogenic responses.
A total HCP extract (cell lysate) specific to a given product should be characterized initially to build a spectral library of potential HCPs. Those within the drug product sample can be identified down to low-ppm levels by comparison with the spectral library.
Product samples throughout product development should be assessed to monitor clearance of HCPs. Once identified, HCPs can be quantitated using a relative quantitative approach by comparison with protein standards added to the sample. More accurate quantitation for specific HCPs of interest can be achieved using synthesized heavy-isotope–labeled peptide standards and MRM experiments.
References
1 Bracewell DG, Francis R, Smales CM. The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control. Biotechnol. Bioeng. 112(9) 2015: 1727–1737; doi:10.1002/bit.25628.
2 United States Food and Drug Administration Code of Federal Regulations, 21 CFR Part 610.13: Purity. US Food and Drug Administration, Rockville, MD, 1 April 2018.
3 USP 39–NF 34 <1132> Residual Host Cell Protein Measurement in Biopharmaceuticals. United States Pharmacopeia–National Formulary. United States Pharmacopeial Convention, Bethesda, MD, 2016.
4 ICH Q6B: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products. International Council on Harmonization of Technical Requirements for Pharmaceuticals for Human Use, Geneva, Switzerland, 1999.
5 Guideline 3AB1A: Production and Quality Control of Medicinal Products Derived By Recombinant DNA Technology. European Medicines Agency, London, UK, 1995.
Jillian Roamer is a senior scientist, and Dr. Victor Sharov is a senior research scientist at BioPharmaSpec; biopharmaspec.com. Christopher Ziegenfuss, Steven Broome, Dr. Richard Easton, and Dr. Andrew Reason contributed to this article.
You May Also Like





