- Sponsored Content
Designing a MAb Purification Process: Based on Bio-Sequential Chromatography
August 10, 2016
Sponsored by Novasep
 In batch-mode chromatography, operating conditions do not allow full use of the total media capacity (1, 2). Bio-Sequential Chromatography (BioSC®) is an open-loop process for the separation of multicomponent solutions (3). The unique column of the batch mode is replaced by a series of smaller columns (from two to six). This design maximizes the use of the media without product loss along the process sequences. The BioSC® Lab automated system is based on the multicolumn chromatography process, designed and provided by Novasep. Studies have been performed to demonstrate the benefits on IgG purity of a downstream process (DSP) monoclonal antibody (MAb) platform with two continuous chromatography steps (4). This note features the outlines of the method used and the qualitative and quantitative results obtained for the product critical quality attributes (CQAs) (Table 2).
In batch-mode chromatography, operating conditions do not allow full use of the total media capacity (1, 2). Bio-Sequential Chromatography (BioSC®) is an open-loop process for the separation of multicomponent solutions (3). The unique column of the batch mode is replaced by a series of smaller columns (from two to six). This design maximizes the use of the media without product loss along the process sequences. The BioSC® Lab automated system is based on the multicolumn chromatography process, designed and provided by Novasep. Studies have been performed to demonstrate the benefits on IgG purity of a downstream process (DSP) monoclonal antibody (MAb) platform with two continuous chromatography steps (4). This note features the outlines of the method used and the qualitative and quantitative results obtained for the product critical quality attributes (CQAs) (Table 2).
Analytical Methods
Purity and concentration were determined using HPLC-SEC.
IgG purity were measured using SDS-PAGE.
Leaching of protein A ligands and level of host cell protein (HCP) were evaluated using commercial ELISA kits.
DNA content was measured using the PicoGreen method.
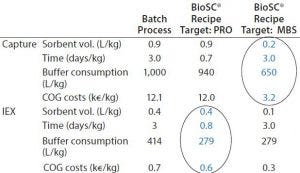
Table 1: Batch and recipe targets generated by BioSC® Predict software
Feed Description: IgG1 subtype MAb was produced from batch cultures of a stable CHO cell line in a 50-L single-use bioreactor.
Resin Screening: Two different protein A media for the capture step (A1 and A2) and 11 ion exchangers for the intermediate step (from IEX1 to IEX11) were evaluated.
Experimental Conditions: For each step, the determination of the optimal process parameters was performed using a laboratory-scale batch column (11-mm ID × 5-cm bed height) by injecting 80% of DBC10%.
The process parameters are
Volume and MAb concentration of the load
Flow rate of the loading step
Buffer concentration
Volumes and pH
Elution buffer conductivity (for ion exchange)
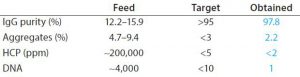
Table 2: Product CQA obtained with BioSC® process
Complete breakthrough curves at optimal conditions were plotted and uploaded in the BioSC® Predict software, for determining the optimized BioSC® recipe for each case. The calculation model can users help reach several objectives depending on the operation target, either the best productivity (PRO scenario) or the reduction of the media and buffer consumption (MBS scenario). Quantitative results show that product CQA can be obtained with two chained BioSC® steps (Tables 1 and 2).
Conclusion
Purification data from MAbs at clinical stage were generated with BioSC® processes. The combination of two successive steps (protein A followed by ion-exchange chromatography) demonstrated a significantly better performance than batch processing (productivity, media and buffer savings) while maintaining product CQAs. Tables 1 and 2 summarize the results.
References
1 Mahajan E, George A, Wolk B. Improving Affinity Chromatography Resin Efficiency Using Semicontinuous Chromatography. J. Chromatog. A 1227, 2012: 154–162.
2 Perez-Almodovar EX, Carta G. IgG Adsorption on a New Protein A Adsorbent Based on Macroporous Hydrophilic Polymers: Pressure-Flow Curves and Optimization for Capture. J. Chromatog. A 1216, 2009: 8348–8354.
3 Holzer M, Osuna-Sanchez H, David L. Multicolumn Chromatography: A New Approach to Relieving Capacity Bottlenecks for Downstream Processing Efficiency. BioProcess Int. 6, 2008: 74–82.
4 Girard V, et al. Large-Scale Monoclonal Antibody Purification by Continuous Chromatography: From Process Design to Scale-up. J. Biotechnol. 10 Nov 2015; 213:65-73. doi: 10.1016/j.jbiotec.2015.04.026.
Hélène Lafontaine is LP product manager, Novasep – Biopharma Business Unit, Site Eiffel BP 50, 81, Boulevard de la Moselle, 54340 Pompey, France; 33-3 83-49-71-47; [email protected]; www.novasep.com.
You May Also Like