Pseudomonas fluorescens: Cell-Line Development of a Commercially Proven Platform for Biopharmaceutical Manufacturing
A number of factors contribute to delivering a robust, highly productive, and reliable process for manufacturing a therapeutic protein. They begin with a cell-host system and a gene-expression strategy that determine a developer’s ability to optimize growth and expression titers. But for many therapeutic proteins, initial attempts to develop a production process are based on evaluation of limited factors and tend to yield only small quantities or poor product quality. Automation can enable parallel building and expression screening of diverse expression strategies within a robust host cell system, accelerating product development. Ensuring significant production of a therapeutic protein from the start, and thus quickly generating high protein quality and yield, can provide significant advantages in opportunity costs.
Formerly known as Pfenex expression technology, the Pelican Expression Technology platform (PET) is a microbial expression system based on the Gram-negative bacterium Pseudomonas fluorescens. The platform was designed to enable rapid, robust, and cost-effective production of proteins, antibodies, and peptides for use in human therapeutics, vaccines, and other applications. With proven advantages in soluble and active protein expression titers, PET reduces production and development costs, enabling rapid delivery of products through preclinical and clinical development and into commercialization. Four commercial products are derived from this validated therapeutic protein production platform (1):
• teriparatide injection (licensed to Alvogen for commercialization in the United States) for osteoporosis
• Pneumococcal conjugate vaccine from the Serum Institute of India
• Recombinant asparaginase erwinia chrysanthemi, a component of a multiagent chemotherapeutic regimen from Jazz Pharmaceuticals
• Pneumococcal 15-valent conjugate vaccine from Merck.
PET Cell-Line Development
The key first step in delivering a robust therapeutic protein production process is to identify an optimal combination of production strain and fermentation condition. The PET group within Ligand Pharmaceuticals uses an innovative approach to strain screening that leverages the unique properties of its proprietary bacterial strain P. fluorescens. We use an automation-enabled, high-throughput (HTP) parallel screening method that can identify an optimized strain rapidly for further development (2, 3). Our strategy takes advantage of the obligate aerobic nature of the P. fluorescens organism to produce enough biomass at 96-well expression scale for generating a target protein in quantities required to complete a reasonable analysis of quantity and quality.
Top-candidate production strains are evaluated further in small-scale fermentations (typically 2-L scale) to assess for productivity, robustness, and protein quality under different induction conditions. Typical parameters tested in a multivariate design of experiments (DoE) approach include temperature, pH, inducer concentration, and cell density at induction. Enrichment and quality assessment of protein expressed at fermentation scale are key to selecting a final production strain. We routinely use automated HTP protein enrichment with miniaturized chromatography columns and/or 96-well resin plates to generate samples for analytical characterization, including for cell-based assays when applicable.
The PET platform is derived from P. fluorescens biovar I MB101 strain, an aerobic Gram-negative bacterium that is a natural, nonpathogenic, and abundant component of the microbial flora in soil, water, and plants. P. fluorescens is metabolically versatile, grows on a wide range of organic substrates, and maintains its viability for long periods in diverse environments. The microbe grows best at neutral pH and 32 °C temperature. It can be cultivated to high cell densities in defined mineral-salts media supplemented with inorganic nitrogen and a carbon source such as glucose or glycerol (4). Because P. fluorescens is a strict (obligate) aerobe, maintaining sufficient oxygen transfer to the cultures promotes higher cell densities with minimal accumulation of organic acids that can be detrimental to growth.
In Escherichia coli, glucose is transported by the phosphoenolpyruvate (PEP)-dependent transport system (PTS), which drives sugar-substrate uptake and phosphorylation in a single energy-coupled step. Pseudomonads lack the PTS system for glucose transport; they take up glucose preferentially through oxidative assimilation rather than through a phosphorylative pathway (5). Biomass levels >100 g/L (dry cell weight) can be achieved at production scales without requiring oxygen supplementation. Optical densities (OD600) of >40 can be attained easily at 96-well growth scale (0.5-mL culture volume) with excess nutrients and high orbital shaking speeds. PET platform cultures use media containing no animal-derived components and no antibiotics at any point in the process. Protein production plasmids are maintained stably through use of auxotrophic complementation as a selection marker (6).
An extensive toolbox of genetic elements and host-strain phenotypes can be screened quickly in parallel with the above parameters to identify combinations that promote high specific expression of soluble and active protein. Based on intrinsic properties such as amino acid sequence, it is difficult to predict a priori which options will express a given protein. Therefore, applying a combinatorial, parallel processing approach to strain screening can improve success and accelerate product development by generating more promising leads and faster go–no-go decisions.
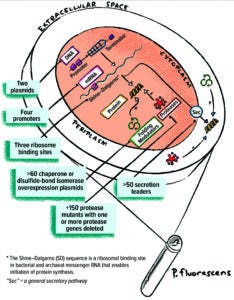
Figure 1: The Pelican Expression Technology (PET) toolbox of genetic elements and host strains.
Pelican toolbox components can be combined to produce thousands of unique expression strains for parallel screening of protein expression (Figure 1). Tools include specific host-strain phenotypes developed to promote soluble and active expression without proteolytic clipping (deletion of one or more chromosomal protease encoding sequences) and co-overexpression of protein-folding helpers such as chaperones and disulfide-bond–formation factors. Many toolbox host strains combine both phenotypes. Additionally, >100 off-the-shelf expression vectors contain unique combinations of incorporated genetic control elements including transcriptional promoters, translation initiation strengths (ribosome binding site (RBS) sequences), and secretion-signal peptide-coding sequences. With a single cloning strategy, genes optimized using different strategies can be ligated into the toolbox expression vectors for subsequent expression screening.
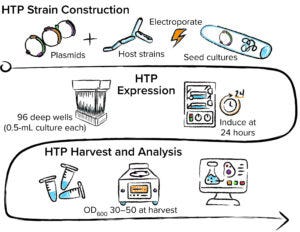
Figure 2: Automation-enabled high-throughput (HTP) strain construction, expression, and analysis performed at 96-well scale.
Those host strains and expression plasmid arrays are transformed, grown, and induced at 96-well scale using automated equipment (Figure 2). Automated workflows also are used to process growth samples for analytical evaluation. Culture samples induced for protein expression typically are sonicated at 96-well scale. The generated lysate is analyzed for protein titer and quality using HTP sodium dodecyl sulfate–capillary gel electrophoresis (SDS-CGE). If a binding partner is available, then biolayer interferometry (BLI) can measure quantities of active binding protein in lysate samples. Titer data are used to identify combinations of expression plasmid strategy and host-strain phenotype that promote soluble and active therapeutic protein. Further quality assessments also can be performed at this scale: e.g., confirming expressed protein intact mass using liquid chromatography with mass spectrometry (LC-MS) or capillary electrophoresis with electrospray ionization MS (CE/ESI-MS).
Using the PET platform, we can identify a robust and productive expression strain within 9–12 weeks from the start of gene cloning to 2-L enrichment, quickly enabling further process development. Hundreds of proteins in a number of classes including antibody derivatives, vaccine antigens, therapeutic enzymes, human cytokines, and growth factors have been expressed using the platform with a success rate of >80% for production of soluble and active proteins that have failed in other systems including yeast, E. coli, insect cells, and Chinese hamster ovary (CHO) cells (Figure 3). In addition, the flexible toolbox can be used to evaluate multiple therapeutic candidates in parallel, enabling rapid transition from lead identification to development for clinical studies.
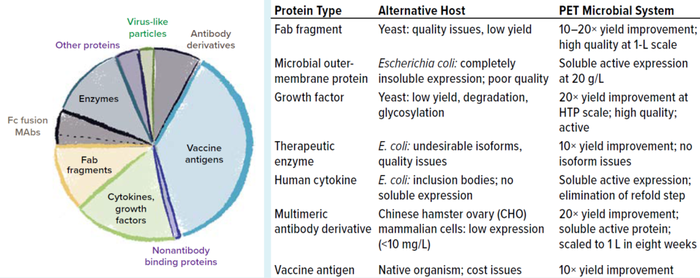
Figure 3: Examples of PET improvements to therapeutic protein expression across hundreds of case studie.
Case Study: Identifying a Strain Candidate
Using PET workflows enabled identification of a strain candidate for a neutralizing recombinant human Fab antibody fragment for continued product development within 12 weeks. A total of 800 expression strains, each reflecting a unique combination of toolbox elements, were evaluated at 96-well scale for Fab expression using automated, HTP workflows. Strains were designed specifically for periplasmic expression of Fab heavy and light chains using secretion peptide fusions incorporated in toolbox plasmids. Periplasmic expression of Fab is desirable to take advantage of the less-reducing environment (compared with that of the cytoplasm) and folding factors that promote Fab heavy- and light-chain assembly. Induced, assembled Fab fragments in culture-soluble lysate fractions were analyzed by HTP SDS-CGE. Down-selected samples showing an induced band by SDS-CGE were analyzed with BLI to quantify titers of recombinantly expressed Fab actively binding to its target. Three strains representing diverse expression plasmids and host strains were chosen for evaluation at 2-L–scale fermentation under eight different induction conditions (Table 1). Those three strains showed assembled Fab titers at 96-well expression scale ranging from 308 mg/L to 352 mg/L by SDS-CGE and 209 mg/L to 219 mg/L by BLI.
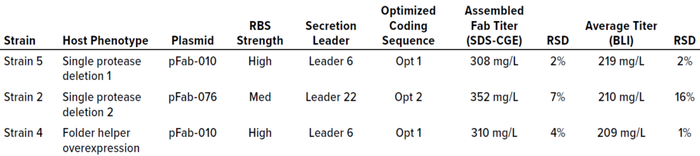
Table 1: Three strains producing the highest titers of active Fab at 96-well scale measured by both sodium dodecyl sulfate capillary gel electrophoresis (SDS-CGE) and biolayer interferometry (BLI) were down-selected for evaluation at fermentation scale. These strains represent a diversity of expression-plasmid strategies — a combination of ribosome binding site (RBS) strength, secretion leader, and optimized coding sequence — and host-strain phenotypes. RSD = relative standard deviation.
For fermentation at 2-L scale, the induction parameters of pH, temperature, biomass density at induction, and inducer concentration were evaluated by fractional factorial design to identify combinations of strain and fermentation conditions yielding the highest quantity of soluble and active Fab product. Culture lysates were analyzed by SDS-CGE (Figure 4) and BLI over the time course postinduction.
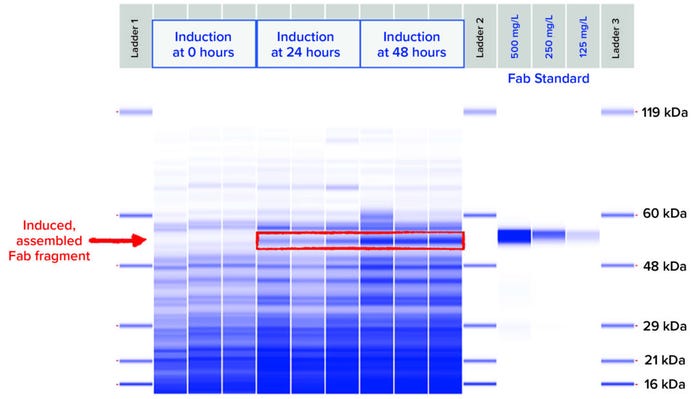
Figure 4: SDS-CGE gel-like images of nonreduced soluble lysate samples from Strain 2 grown and induced at 2-L scale under the most productive induction condition.
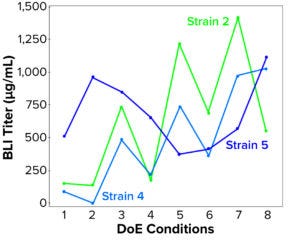
Figure 5: Soluble lysate BLI titer (mcg/mL) compared with design of experiments (DoE) treatment number for three strains grown and induced at 2-L–scale strain scouting
We screened the three scaled strains efficiently in parallel to investigate induction-parameter combination effects on performance and identify key fermentation control parameters. Strain 5 showed a different pattern in its response to the parameters tested than those of Strains 2 and 4, suggesting that a different approach should be taken to optimize and control the latter two (Figure 5).
By testing the three strains across a number of parameters, we quickly identified the Fab expression strain (Strain 2) that exhibited highest productivity over a range of conditions. Furthermore, we explored in more detail the induction parameters for Strain 2 that had the largest impact on protein titer and quality and refined the duration of induction to develop the Fab production process further. The robustness of the production host enabled extension of the production phase to at least 96 hours. During this process, the soluble active Fab titer improved from 0.21 g/L at the HTP scale to 4.8 g/L for the final process at 2-L scale (Table 2). Samples enriched from fermentation paste by affinity chromatography were confirmed for active binding (by BLI) and correct mass (by LC-MS) of assembled Fab.

Table 2: Strain 2 soluble active Fab titers measured by biolayer interferometry (BLI) during high-throughput (HTP) screening and 2-L fermentation evaluation, then after 2-L process development.
Strain 2 uses different Pelican toolbox secretion leaders for localization of the Fab heavy- and light-chain subunits to the periplasmic space of cells to enhance disulfide bond formation and folding. Additionally, the heavy-chain fragment required a medium translational initiation strength (RBS sequence) whereas the light-chain fragment required a high-translation initiation strength to achieve an optimal ratio of the Fab subunits to promote efficient assembly into the active form. Strain 2 contains a deletion of a specific protease to alleviate target protein degradation.
Identifying Optimal Expression Strategies
The PET team has developed a powerful protein expression technology based on P. fluorescens that leverages an extensive combinatorial toolbox and automated workflows. The platform can quickly identify optimal expression strategies to support therapeutic protein development. To date, four products have been approved for major pharmaceutical developers using this platform. Key to its performance is rapid and robust protein analysis using HTP titer assays (including SDS-CGE and BLI technologies) to screen thousands of strains in parallel at 96-well scale in a fully automated system. Highly productive strains identified at 96-well scale are advanced to fermentation scale and scouted under more controlled growth and induction conditions using a DoE strategy to develop manufacturing strains. Additionally, the PET platform has been successful in late-stage discovery studies to enable hit-to-lead selection of antibody derivatives, mammalian cytokines, growth factors, enzymes, and viral and microbial proteins.
References
1 Pelican Expression Technology: Streamlining Protein Therapeutic Development. March 2022; https://pelicanexpression.com.
2 Retallack DM, et al. Transport of Heterologous Proteins to the Periplasmic Space of Pseudomonas fluorescens Using a Variety of Native Signal Sequences. Biotechnol. Lett. 29(10) 2007: 1483–1491; https://doi.org/10.1007/s10529-007-9415-5.
3 Retallack DM, Jin H, Chew L. Reliable Protein Production in a Pseudomonas fluorescens Expression System. Protein Expr. Purif. 81(2) 2012: 157–165; https://doi.org/10.1016/j.pep.2011.09.010.
4 Production of Recombinant Proteins: Novel Microbial and Eukaryotic Expression Systems (1st ed). Gellissen G, Ed. Wiley-VCH Verlag: Weinheim, Germany, 2006.
5 Temple LM, et al. Carbohydrate Catabolism in Pseudomonas aeruginosa. Biotechnology Handbooks 10: Pseudomonas. Springer US: Boston, MA, 1998: 35–72.
6 Schneider JC, et al. Auxotrophic Markers pyrF and proC Can Replace Antibiotic Markers on Protein Production Plasmids in High-Cell-Density Pseudomonas fluorescens Fermentation. Biotechnol. Prog. 21(2) 2005: 343–348; https://doi.org/10.1021/bp049696g.
Further Reading
Espinosa DA, et al. The Plasmodium falciparum Cell-Traversal Protein for Ookinetes and Sporozoites as a Candidate for Preerythrocytic and Transmission-Blocking Vaccines. Infect. Immun. 85(2) 2017: e00498-16; https://doi.org/10.1128/iai.00498-16.
Maese L, et al. Can Recombinant Technology Address Asparaginase Erwinia chrysanthemi Shortages? Pediatr. Blood Cancer68(10) 2021:e29169; https://doi.org/10.1002/pbc.29169.
Noe AR, et al. A Full-Length Plasmodium falciparum Recombinant Circumsporozoite Protein Expressed By Pseudomonas fluorescens Platform as a Malaria Vaccine Candidate. PLoS One 9(9) 2014: e107764; https://doi.org/10.1371/journal.pone.0107764.
Reed MD, et al. Immunization with a Recombinant, Pseudomonas fluorescens-Expressed, Mutant Form of Bacillus anthracis-Derived Protective Antigen Protects Rabbits from Anthrax Infection. PLoS One 10(7) 2015: e0130952; https://doi.org/10.1371/journal.pone.0130952.
Schneider JC, et al. Safety and Immunogenicity of Px563L, a Recombinant Anthrax Vaccine Candidate, in a Two-Dose Regimen for Post-Exposure Prophylaxis in Healthy Adults. Vaccine 39(42) 2021: 6333–6339; https://doi.org/10.1016/j.vaccine.2021.08.075.
Corresponding author Russell Coleman is director of strain engineering, Elizabeth Orchard is fermentation group lead, and Yinghui Lee is a senior scientist at Ligand Pharmaceuticals, 10790 Roselle Street, San Diego, CA 92121; 1-858-550-7500; https://www.ligand.com. Pelican Expression Technology (PET) is a registered trademark of Ligand Pharmaceuticals.
You May Also Like





