
As demands increase for antibody therapeutics from both patients and regulators, new modalities arise that require critical analysis and multiattribute methods. This month's Featured Report highlights developments in antibody manufacturing, from artificial intelligence (AI)-assisted protein engineering to production, purification, and characterization.
Intact and Subunit Molecular Mass Analysis for Development of Antibody Therapeutics: Considerations for Novel Biotherapeutics and BiosimilarsIntact and Subunit Molecular Mass Analysis for Development of Antibody Therapeutics: Considerations for Novel Biotherapeutics and Biosimilars
Biologics play a vital role in the pharmaceutical sector and hold much promise for improving health. Approved protein drugs and biosimilars are making their largest impacts in the oncology and autoimmune domains. Monoclonal antibodies (mAbs) form the largest segment of biopharmaceuticals with complex molecular manifestations such as posttranslational modifications (PTMs). Recent approvals for bispecific antibodies (bsAbs) and antigen-binding fragments (Fabs) now highlight the importance of analytical characterization for such multifaceted entities.
Development of antibody-based therapeutics demands cutting-edge technology to characterize their critical quality attributes (CQAs) (1). At early stages, intact and subunit molecular-mass analyses can provide quick and accurate glances at the CQAs of biotherapeutics undergoing development or biosimilarity assessment. Several strategies have been used to measure protein molecular mass at both intact and subunit levels, with a focus on variants. Early stage inputs to antibody/biosimilar development can ease feasibility assessments, in turn providing confidence to drug manufacturers (2). Compared with early analyses, late-stage monitoring tends to be much more straightforward because it involves prequalified/validated methods that are directly applicable to batch analysis and release. Early characterization activities also support chemistry, manufacturing, and controls (CMC) teams with comprehensive data packages that help to ensure drug program success at all stages.
Antibody products require molecular-mass measurement at both the intact and subunit levels, providing a plethora of analytical information for product understanding. Complexities from glycosylation, terminal lysines, and many such modifications account for much of the difficulty in characterizing an entire molecule. Similar approaches may be directed toward bsAbs and antibody–drug conjugates (ADCs), with analytical emphasis on heavy-chain variants and drug:antibody ratios (DARs), respectively.
By leveraging continually improving sample-preparation strategies, ultraperformance liquid chromatography (UPLC) and high-resolution mass spectrometry (HRMS) now lead efforts to obtain critical information about molecular mass. Multiattribute methods (MAMs) for intact and subunit analysis also can provide rapid and accurate monitoring platforms for accelerated biotherapeutic development and manufacturing — not to mention separation methods. Reversed-phase (RP) high-performance liquid chromatography (HPLC), size-exclusion chromatography (SEC), ion-exchange chromatography (IEC), hydrophilic-interaction chromatography (HILIC), hydrophobic-interaction chromatography (HIC), and migration by capillary electrophoresis (CE) all can be integrated with HRMS (3). Below, we explore applications of such methods to CQA evaluation for antibody-based therapeutics, including novel biological entities (NBEs) and biosimilars.
Importance of Measuring Molecular Mass
Molecular-mass measurements provide a quick and accurate way of confirming a protein’s primary structure. The resulting data provide a comprehensive view of an entire protein built on amino-acid sequences and associated modifications (e.g., glycosylation patterns). Because of heterogeneity in proteoglycan–lysine forms, strategies based on improved sample preparation, robust separation, and HRMS are needed to provide reliable and interpretable data. Methods are available for selecting specific forms based on coordinated application of suitable reagents and enzymes. Such approaches can offer deep insights when investigating associated glycoforms, evaluating clearance of lysine variants, and detecting previously unknown variants.
A “step-down” approach to studying proteins in their subunit forms has proved to be a great advantage for antibody-based biotherapeutics. Each subunit provides critical information about a protein’s structural and functional integrity. Analysis of heavy chains accounts for the most important aspect of antibody specificity: glycosylation. Heavy chains are also critical sites for monitoring charge variants (e.g., C-terminal lysines). Light chains provide diversification in molecular binding sites, so primary-structure characterization is essential in that context. Detailed insights also can come from subunit studies based on enzymatic digestions separating the Fab and crystallizable fragment (Fc) portions.
Studies based on those approaches will provide comprehensive understanding about a protein’s backbone and glycosylation pattern as well as the presence or absence of terminal lysines and unanticipated forms. For targeted digestion, structural analyses leverage enzymes such as glycosidase, carboxypeptidase, and cysteine proteases. Strategic application of such enzymes in combination with agents for reduction/alkylation can generate multiple subunit products for molecular investigation.
Typical mAbs average ~150 kDa in mass, with glycoforms accounting for mass differences of~162 Da. Such variants can be attributed, for instance, to additions of galactose to a core glycan structure. Apart from glycoforms, terminal lysines present on the C-terminals of heavy chains can create antibody species with a difference of ~128 Da. In some (albeit unlikely) situations, we might expect to find modifications such as oxidation, sequence variants, and unidentified species. All of the above modifications contribute to the delta mass changes that become apparent during molecular-mass analyses. Treating antibody samples with specific enzymes can provide the selectivity and analytical clarity needed to distinguish glycoforms, lysine forms, and other variants from the protein backbone.
Introducing reduction and/or alkylation agents in combination with enzymes can generate multiple subunits for study, including heavy chains, light chains, Fabs, Fc regions, and Fab heavy chains (Fd regions). Laboratories for both early drug-development stages and routine monitoring require robust methods to investigate such species for changes in structural integrity. Being a mediator of biological activity, an antibody’s Fc region can provide information related to hotspots for glycosylation and terminal lysines.
Enzymatic strategies similar to those used for intact-protein analyses can be extended to study Fc regions in detail. On the other hand, characterizing light chains, Fd regions, and Fabs reveals information about antigen binding, which occurs in an antibody’s complementarity-determining regions (CDRs).
Table 1 lists methodologies for analysis of intact and subunit molecular mass of proteoglycan forms and associated variants. The table also describes what sample-preparation strategies are needed to generate particular entities for investigation. For example, intact protein analysis using peptide N-glycosidase F digestion (InProP) removes glycans from intact proteins, providing molecular mass for their backbones and lysine forms (if any). By contrast, intact analysis based on N-glycosidase F and carboxypeptidase digestion (InProPC) cleaves glycans and terminal lysine, enabling analysts to derive the molecular mass of a protein backbone. Figure 1 depicts the applicability of such sample-preparation methods to the top three categories of antibody therapeutics in the oncology space: mAbs, bsAbs, and ADCs. Methods for analysis of heavy-chain variants and DARs apply to bispecific and ADC characterization, respectively.
Figure 1: Characterization methodologies and their applicability to monoclonal antibodies (mAbs), bispecific antibodies (bsAbs), and antibody–drug conjugates (ADCs)
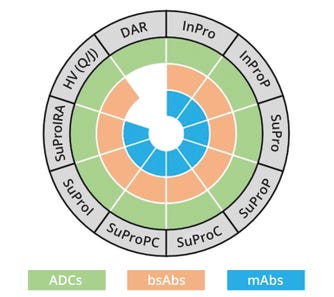
DAR = analysis of drug:antibody ratio, HV (Q/J) = analysis of homologous/heterologous heavy chains from a bispecific antibody, with Q and J representing each chain, InPro = analysis of intact protein, SuPro = analysis of protein subunit; A = alkylation, C = digestion by carboxypeptidase B, I = digestion by immunoglobulin-G–degrading enzyme of Streptococcus pyogenes (IdeS), LC = light chain, P = peptide N-glycosidase F, R = reduction
Leveraging Molecular Features for Separation Processes
Antibodies are combinations of amino acids with associated PTMs. Such features present opportunities to extend characterization. Antibodies are known to be expressed in various proteoforms, sizes, and charges. Rather than relying on one methodology, analysts should leverage multiple separation chemistries, with principles based on differences in hydrophobicity, hydrophilicity, size, and/or charge.
Analytical laboratories equipped with state-of-the art technologies will be fertile ground for exploring applicability of CE, IEC, HIC, HILIC, RP-HPLC, and SEC as well as their connectivity with MS. A particularly important consideration is technical competence in selecting suitable methods (e.g., separation chemistries) and materials (columns, reagents, and enzymes). Analysts also must perform due diligence to ensure proper handling of processed samples.
Understanding process- and product-related variants is critically important when submitting applications for regulatory review of NBEs and biosimilars. Developers should anticipate the need for extensive analytical programs for variant identification and characterization. Applying diverse analytical methods often is a suitable way forward because of variant complexity. Available separation chemistries can be leveraged to develop and establish platform methods for different biotherapeutic classes.
The “Separation Methods” box overviews available separation technologies that can be leveraged for detection, identification, and characterization of product variants in biotherapeutics. Such methods can be integrated with MS to bring in additional data for verification. Two-dimensional (2D) chromatography could be applied in complex situations that previously have called for multimodal separation methods such as RP-SEC, RP-IEC, and SEC-IEC.
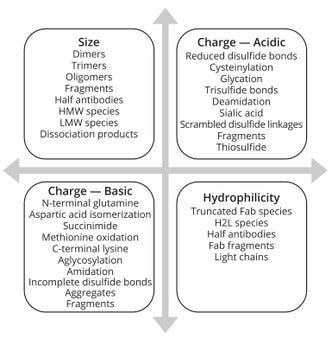
Figure 2 shows multiple dimensions for selecting a suitable approach to variant identification and characterization. Variants in charge, size, and hydrophilicity/hydrophobicity necessitate focused efforts to harness viable methodologies. Complementary data from MS can provide additional support to such analytical information.
Figure 2: Variants observed in mAb and other protein-based products; Fab = antigen-binding fragment, H2L = variant with two heavy chains and one light chain, HMW = high molecular weight, LMW = low molecular weight
Enhancing Capabilities in Molecular-Mass Determination
In the world of therapeutic antibodies, molecular mass is a key indicator of structural integrity. But antibody components span a wide range of masses. Entities such as variable domains (e.g., variable light and heavy chains, VL and VH) can have a mass of~15 kDa, whereas immunoglobulin new antigen receptor (IgNAR) antibodies derived from sharks can reach up to ~175 kDa in their monomeric state. Moreover, fusion proteins such as etanercept and aflibercept pose difficulties for measuring mass, requiring domain expertise and supportive technologies. Native MS is one such solution that can help to resolve molecular heterogeneity and thereby deduce meaningful MS data.
Figure 3 provides a holistic view of molecular-weight ranges for different entities that might be observed during intact-protein and subunit mass analyses. That range typically spans from ~20 Da to 180 kDa, with oligomers reaching as high as ~600 kDa. Analysts generally should focus on that range when developing and fine-tuning their methods. On the other hand, oligomers and aggregates might be cause for increasing the measuring range, so suitable — MS-compatible — methods need to be explored. Given such analytical challenges, it is essential to connect with an expert team that leverages proven technologies.
Figure 3: Range of molecular masses for intact and subunit proteins/antibodies
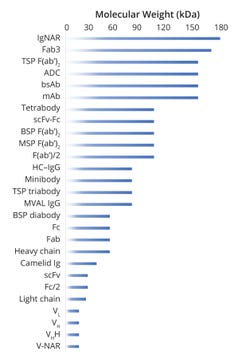
ADC = antibody–drug conjugate, bsAb = bispecific antibody,
BSP = bispecific, Fab = antigen-binding fragment, F(ab’)2 = fully intact Fab, F(ab’)/2 = Fab fragment, Fab3 = Fab fragments from trivalent antibody, Fc = crystallizable fragment region, Fc/2 = Fc fragment,
HC–IgG = human protein HC–immunoglobulin G complex, Ig = immunoglobulin, IgNAR = immunoglobulin with new antigen receptor, mAb = monoclonal antibody, MSP = monospecific, MVAL = monovalent, scFv = single-chain fragment variable, TSP = trispecific, VH= heavy-chain variable domain, VHH = Fab of heavy-chain–only antibody,
VL = light-chain variable domain, V-NAR = variable domain from IgNAR
From selection of separation chemistries to MS execution, analysts must make coordinated efforts to develop mass-analysis methods that will return results with suitable accuracy and precision. Analysts must identify chromatography columns with the right dimensions and conditions so that molecules are resolved according to a quality by design (QbD) gradient method. Mass spectrometers must be fine-tuned to ionize, separate, and detect large molecular entities, helping to ensure good quality in raw data. Thorough evaluation of raw data is critical to monitoring mass:charge (m/z) distributions, ionization patterns, intensities, and related parameters. After evaluation, raw data must be processed for identification, annotation, and interpretation using a robust informatics platform with necessary data-integrity compliances in place. Options for custom fields and report designs can add value to user experiences while providing data with statistical and scientific significance. Systems for digital data review can streamline approval processes while providing for data integrity throughout the measurement journey.
The Balancing Act of Biosimilar Characterization
Contract research organizations (CROs), contract development and manufacturing organizations (CDMOs), and other service providers with strong holds on analytical and characterization capabilities support the ecosystem for investigating NBEs. Such organizations can extend similar setups and capabilities for biosimilarity assessments. Innovations that drive NBE research and development (R&D) also can provide deep insights into aspects of biosimilarity. That said, adequate tools for comparing biosimilars with reference medicinal products (RMPs) are both add-ons and must-haves for evaluation.
Aside from methods for demonstrating comparability between RMPs and candidate biosimilars, powerful informatics platforms are the need of the hour. Such technologies help to generate mirror plots, calculate relative percentages of proteoforms and variants, and so on. Analytics platforms must be implemented according to a compliance-driven approach that integrates processing-method development with data analysis, interpretation, review, and reporting.
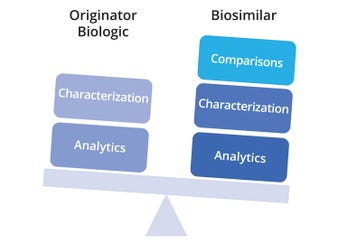
From an analytical perspective, reference biologics and biosimilars require similar platforms and methods, but biosimilars demand comparative outputs as an additional component (Figure 4). Comparative characterization is key to the totality of evidence that must be presented for biosimilar approval, and that factor plays a major role in slowing or expediting the process. The biosimilar route depends heavily on a well-defined analytical package comprising quality target-product profiles (QTPPs), mapped CQAs, risk assessments, design spaces, and control strategies.
Figure 4: Analytical efforts for originator and biosimilar biologics, with the differentiating factor of comparability assessment
Although process-development laboratories offer flexibility in exploring analytical parameters to provide robust methods, it is essential to draw upon lessons from compliance-driven manufacturing environments. For instance, laboratories should implement data-integrity principles, including ALCOA++ standards (for for data attributability, legibility, contemporaneity, originality, accuracy, completion, consistency, endurance, availability, and traceability). Related measures include user-license–based logins, audit-trail–enabled data acquisition and processing systems, and provisions for electronic-record signatures. Adapting to analytical guidelines from the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), United States Food and Drug Administration (FDA), European Medicines Agency (EMA), and other global health authorities will help analytical laboratories to keep abreast with regulatory guidances, opinions, and updates (5, 6).
Biosimilar-product characterization typically involves thorough collection of analytical data from LC-MS, which can generate comparative profiles for a target (biosimilar) and reference product. Used either as stand-alone methods or in tandem, LC and MS must be applied under a compliance-driven approach for data acquisition, processing, and interpretation (6).
Chromatography data systems and MS software platforms with the compliance features noted above are preferred to protect the integrity of analytical data, which form the basis of biosimilar data packages (7). Ultimately, laboratories must be equipped with advanced and regulatory-compliant analytical technologies to provide solutions to the complex challenges of biotherapeutic characterization.
Final Considerations
Molecular-mass measurements are important attributes for antibody-based therapeutics because such drugs comprise complex features such as chains, fragments, and interlinkages. In the case of ADCs, complexities also stem from characterizing conjugation sites. The criticality of such information is acknowledged in regulatory guidelines, and characterization data from analytical packages are reviewed carefully. Leveraging chromatographic separation principles coupled with MS has empowered molecular characterization workflows to solve difficult analytical problems. Similar approaches can be extrapolated to biosimilars with more emphasis on comparative assessment.
Recent discussions among regulators, industry experts, and key opinion leaders (KOLs) highlight the importance of MS-led methodologies in biopharmaceutical characterization. A solution partner that can provide comprehensive, streamlined, end-to-end analytical solutions will help to deliver excellent results. As evidenced by the latest RMP and biosimilar approvals, analytical information primarily from MS has been instrumental in regulators’ consideration of product applications (4).
References
1 Geigert J. The Challenge of CMC Regulatory Compliance for Biopharmaceuticals (Fourth Edition). Springer: Cham, Switzerland, 2023; https://doi.org/10.1007/978-3-031-31909-9.
2 Auclair J, Rathore AS. Intact Mass Analysis of Biopharmaceuticals as Its Own Unique Application. LCGC North Amer. 39(11) 2021: 548–553; https://www.chromatographyonline.com/view/intact-mass-analysis-of-biopharmaceuticals-as-its-own-unique-application.
3 Staub A, et al. Intact Protein Analysis in the Biopharmaceutical Field. J. Pharm. Biomed. Anal. 55(4) 2011: 810–822; https://doi.org/10.1016/j.jpba.2011.01.031.
4 Mans J, et al. The Use of Mass Spectrometry in Therapeutic Protein Biologics License Applications: A Retrospective Review Revisited. J. Am. Soc. Mass Spec. 34(11) 2023: 2575–2584; https://doi.org/10.1021/jasms.3c00286.
5 Vessely C, Bussineau C. QbD in Biopharmaceutical Manufacturing and Biosimilar Development. Biosimilars: Regulatory, Clinical, and Biopharmaceutical Development. Gutka HJ, Yang H, Kakar S, Eds. Springer: Cham, Switzerland, 2018, 187–219; https://doi.org/10.1007/978-3-319-99680-6.
6 Vandekerckhove K, Reeve R. Principles of Analytical Similarity Assessment. Biosimilars: Regulatory, Clinical, and Biopharmaceutical Development. Gutka HJ, Yang H,
Kakar S, Eds. Springer: Cham, Switzerland, 2018, 261–303; https://doi.org/10.1007/978-3-319-99680-6.
7 Sullivan PM, DiGrazia LM. Analytic Characterization of Biosimilars. Amer. J. Health Sys. Pharm. 74(8) 2017: 568–579; https://doi.org/10.2146/ajhp150971.
Further Reading
Kaltashov IA, Wang S, Wang G. Mass Spectrometry in Biopharmaceutical Analysis. Walter de Gruyter GmbH: Boston, MA, 2022; https://doi.org/10.1515/9783110546187.
Rathore AS, Mhatre R, Eds. Quality by Design for Biopharmaceuticals: Principles and Case Studies. John Wiley & Sons: Hoboken, NJ, 2011.
Houde DJ, Berkowitz SA, Eds. Biophysical Characterization of Proteins in Developing Biopharmaceuticals. Elsevier: Amsterdam, Netherlands, 2019.
Kaur H, Reusch D, Eds. Monoclonal Antibodies: Physicochemical Analysis. Elsevier: Amsterdam, Netherlands, 2021.
Rajiv Bharadwaj, PhD, is head of analytics and characterization, and Sanjib Banerjee, PhD, is chief operating officer, both in the Biopharma Division of Veeda Clinical Research Ltd., No. P-3 & 3A, First Main Road, Peenya Industrial Area, First Stage, Bengaluru 560058, Karnataka, India.
You May Also Like





