Cell-line development traditionally has focused on genetic engineering of chromosomal DNA in cellular nuclei. Combining technological advances such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) with ever-increasing genomic knowledge has enabled scientists to get impressive performance from microbial, plant, and animal cells. But few researchers have considered the potential for manipulating how genes are transcribed as an avenue for increasing productivity — until recently, that is.
Niall Barron is a professor of biochemical engineering at Ireland’s National Institute for Bioprocessing Research and Training (NIBRT) and at University College Dublin. He earned his PhD in microbial science from Ulster University and his bachelor of science in microbiology from Trinity College Dublin. And he has been a member of the European Society for Animal Cell Technology’s (ESACT’s) executive committee since 2014. Barron presented at two BPI conferences in 2022 on the potential for using epigenetics in biopharmaceutical production cell-line development, and I spoke with him recently about the subject.
Our Conversation
I think of epigenetics as studying the relationships between environmental/behavioral factors and genetic expression. How does this concept translate to cells in culture? We’ve been working on a niche area of epigenetics referred to as epitranscriptomics. That’s epigenetic modifications to RNA rather than DNA. We look at methylation of RNA — protein-encoding mRNA, in particular. The specific type of methylation we’re interested is “m6A,” which is a methyl group added to the sixth nitrogen of the adenosine base.
This is something we’ve known about for quite some time, but elucidating the role of this m6A mark on RNA has become topical in life sciences over the past five or six years. Our interest is whether this particular modification can be useful in the context of producing biological medicines in eukaryotic cells. We don’t use environmental modification to influence that. A paper we published recently describes some of the work we’ve done (1). It’s a genetic approach to influencing m6A on RNA transcripts.
How does that differ from other genetic-engineering approaches? We’re pursuing a novel element of genetic control. There are many layers to how cells and organisms can fine-tune the control of the expression of a particular gene. You have transcriptional control, for example. And you have epigenetic control in the traditional sense using methylation or acetylation of DNA to prevent gene expression.
But cells have a group of methylases and demethylases that can add this m6A mark onto RNA. And then a group of reader proteins dictates the fate of the RNA based on the presence or absence of m6A marks on that particular transcript.
Is this kind of work new in biopharmaceutical development? What technological advances have made it more applicable now than in the past? It’s pretty complex stuff, and we are coming at it from a fairly applied angle. Can this process be manipulated to influence how effectively production cells can make a product in culture? It’s a reasonably new approach. I believe we are the first to publish on using this approach in bioproduction systems.
This epigenetic modification on mRNA is known to influence and play a role in fundamental processes such as development, but also in diseases like cancer and obesity. We’re simply looking at its role through the lens of bioproduction.
From the Most Recently Published Work |
In a 2019 BPI Europe talk on this topic, you said, “Manipulation of mRNA epigenetics represents a potential opportunity to manipulate the [Chinese hamster ovary] CHO production platform, but better understanding will be required first.” How has the general understanding advanced since then? It’s improving but it’s clear that these processes are very complex. The biomedical literature on the role of m6A in influencing cancer, for example, or different disease states seems to be context specific. The presence or absence of an m6A residue in a particular sequence depends on the transcript. It depends on location and other cellular contexts. There have been conflicting reports of particular proteins involved in methylation of RNA, both negatively and positively correlating, for example, to a particular cancer phenotype.
Controlling posttranscriptional regulation of gene expression is challenging. We’ve worked on micro-RNAs for a number of years in the CHO cell field. They affect the expression of potentially many different transcripts. Multiple mRNAs in cells can be affected by an individual micro-RNA, so it’s complicated to figure out the extent of the pathways that manipulation of an individual micro-RNA can affect. And the same seems to go for these methylation markers: It’s a complex regulatory layer of gene-expression control. Everybody’s trying to come to terms with that process and understand it better, both in the context of disease and, in our case, whether it can be used as a tool for influencing production in CHO and other cell types.
What’s the significance of those structural changes in recombinant protein expression? Are they applicable only to mammalian cell work, or also to other expression systems? I mentioned that it’s sequence-specific. And we’ve only studied it in mammalian cell lines — in CHO cells and HEK293 [human embryonic kidney] cells. We’ve investigated the role that m6A plays in CHO-cell production of recombinant proteins and in HEK293 production of lentiviral vectors. Those viruses are used to deliver gene therapies, such as in [chimeric antigen receptor] CAR T cells. That’s the context for our interest there.
Can you briefly describe how the technology works and at what scales it’s been applied successfully? We’ve published two different approaches. The first approach is placing individual DNA sequences in an expression construct, a plasmid that ultimately also encodes for your product of interest (2). Human erythropoietin (Epo) is one example. By placing a specific sequence (containing an A residue that’s an m6A target) upstream of the coding sequence, we improved production of Epo in subsequently transfected CHO cells. Naturally occurring m6A sites also influence the expression of protein from local coding sequences, such as one found in the 5’UTR of the mouse Hsp70 gene (Figure 1).
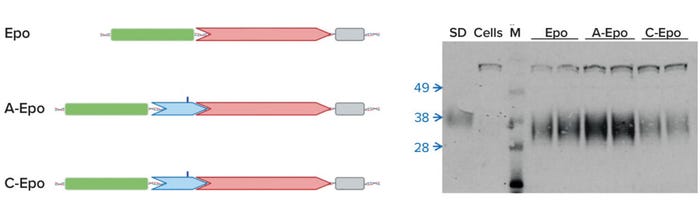
Figure 1: The presence of m6A in the 5′UTR of mouse Hsp70 enhances translation; Epo = erythropoietin (no UTR), A-Epo = wild-type 5′UTR, C-Epo = mutated 5′UTR with m6A changed to C.
Our second approach is to influence the machinery within a production cell line, specifically those reader proteins that bind to m6A-containing transcripts and influence their fate (1, 3). We used siRNA to downregulate a specific reader called YTHDF2 in CHO cells to, more or less, double the yield of Epo (as a model therapeutic gene) from those CHO cells.
A similar approach — downregulation of the YTHDF2 protein — in HEK293 can more or less double the production of lentiviral vectors with standard triple or quadruple transfection of plasmids into those cells. We’ve done this at small scale
(10–20 mL) only just to prove the concept. Whether it scales remains to be seen.
Having seen the yield benefits of knocking down that particular protein, we plan on deleting that gene completely either from CHO or HEK cells. Doing so could help us to determine whether we can generate a new host-cell line for use in recombinant protein production at commercial scales. What we’ve done so far is all transient work at small scale.
Do you use any similar techniques to those used in mRNA therapeutics development? We have a project at the moment involving mRNA therapeutics. We’ve shown that manipulating the presence or absence of m6A residues in certain transcripts can influence their fate (by increasing their stability, in particular). And obviously, there’s a lot of interest in the biopharmaceutical industry for increasing mRNA stability inside cells. This is different from product stability during storage, transport, and delivery, which we’ve all become aware of subsequent to the pandemic.
But mRNA half-life inside cells is relevant to vaccine production and to using mRNA as a vehicle for therapeutic-protein administration. Once you administer a vaccine or therapeutic dose, you want that mRNA to remain intact in a recipient’s cells for as long as possible to make your vaccine or therapeutic protein.
We are interested currently in finding a way to use an in vitro methylation approach to add methyl groups to specific adenosines. But it must be targeted. You can’t just randomly incorporate methylated adenosines into a transcript and thus increase its stability. In fact, it would have the opposite effect. You get little or no translation from randomly methylated transcripts. So we’re working on a more targeted approach.
That brings up the possibility of using CRISPR [clustered regularly interspaced short palindromic repeats] as a vehicle. CRISPR with Cas9 [CRISPR-associated protein 9] has been used for targeted double-strand breaks. The “dead Cas9” (dCas9) approach uses the protein and a guide to locate that complex at a particular sequence in the genome. There are Cas proteins that bind to RNA rather than DNA. An interesting publication from David Liu’s laboratory at the Broad Institute a couple of years ago described a dCas9 fused to a methylase enzyme, and the authors demonstrated targeted methylation of transcripts in vivo (4).
We’re interested in taking essentially that construct — a Cas fused to the methylase — and seeing whether we can achieve the same thing in vitro with a view to generating targeted methylation events on [in vitro transcription] IVT-generated RNA that could be used for subsequent applications. One of those would be generating CAR T cells.
Are there any drawbacks that must be balanced against the benefits? The complexity of influencing methylation within a cell en masse could have potential downsides, but those are yet to be determined. Our next steps include trying to improve some targeted approaches for adding specific methyl groups in particular places or indeed ablating them to benefit somehow. You can increase translation, improve stability, and get other mechanistic effects by influencing those methylation markers.
Do epigenetic approaches provide orthogonal techniques that can be applied alongside (or after) advanced gene-modification work using CRISPR, for example? Are they two sides of the same coin, so to speak? Yes, you can either combine CRISPR approaches or use them subsequently. You could make genetic modifications and then epigenetic modifications on the resulting RNA.
Outside of your work, what are some epigenetics applications that have intrigued you? At David Liu’s laboratory, they are combining the localization capability of a CRISPR approach with targeted methylation (4). I think that’s a really interesting way to think about it for the future.
References
1 Lao N, Barron N. Enhancing Recombinant Protein and Viral Vector Production in Mammalian Cells By Targeting the YTHDF readers of N6-Methyladenosine in mRNA. Biotechnol. J. 24 January 2023: 2200451; https://doi.org/10.1002/biot.202200451.
2 Lao N, Barron N, Clynes M. Continuous Translation of Circularized mRNA Improves Recombinant Protein Titer. Metabol. Eng. 52, March 2019: 284–292; https://doi.org/10.1002/biot.202200451.
3 Lao N, Barron N. Cross-Talk Between m6A and m1A Regulators, YTHDF2, and ALKBH3 Fine-Tunes mRNA Expression. bioRxiv 26 March 2019; https://doi org/10.1101/589747.
4 Rees HA, Liu DR. Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nat. Rev. Genet. 19(12) 2018: 770–788; https://doi.org/10.1038/s41576-018-0059-1.
Further Reading
Bryan L, et al. Differential Expression of miRNAs and Functional Role of miR-200a in High and Low Productivity CHO Cells Expressing an Fc Fusion Protein. Biotechnol. Lett. 43, 2021: 1551–1563; https://doi.org/10.1007/s10529-021-03153-7.
Costello A, et al. Reinventing the Wheel: Synthetic Circular RNAs for Mammalian Cell Engineering. Trends Biotechnol. 38(2) 2020: 217–230; https://doi.org/10.1016/j.tibtech.2019.07.008.
Dhiman H, et al. Genetic and Epigenetic Variation Across Genes Involved in Energy Metabolism and Mitochondria of Chinese Hamster Ovary Cell Lines. Biotechnol. J. 14(7) 2019: 1800681; https://doi.org/10.1002/biot.201800681.
Donohue N, et al. Bio-Production of Adeno-Associated Virus for Gene Therapy. Cell Culture Engineering and Technology. Pörtner R, Ed. Springer: New Yor, NY, 20 February 2022; 335–364; https://doi.org/10.1007/978-3-030-79871-0_11.
Kozak M. Influences of mRNA Secondary Structure on Initiation By Eukaryotic Ribosomes. Proc. Nat. Acad. Sci. USA 83(9) 1986: 2850–2854; https://doi.org/10.1073/pnas.83.9.2850.
Liu N, et al. N6-Methyladenosine–Dependent RNA Structural Switches Regulate RNA–Protein Interactions. Nature 518(7540) 2015: 560–564; https://doi.org/10.1038%2Fnature14234.
Wang X, et al. N6-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161(6): 1388–1399; https://doi.org/10.1016%2Fj.cell.2015.05.014.
Cheryl Scott is cofounder and senior technical editor of BioProcess International (part of Informa Connect Life Sciences); [email protected].
You May Also Like


.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=700&auto=webp&quality=80&disable=upscale)