Voices of Biotech
Podcast: MilliporeSigma says education vital to creating unbreakable chain for sustainability
MilliporeSigma discusses the importance of people, education, and the benefits of embracing discomfort to bolster sustainability efforts.
Monoclonal antibodies (MAbs) are the fastest growing segment in the biopharmaceutical industry because they are potentially efficacious in the treatment of diseases such as cancer and autoimmune disorders (1,2). With steadily increasing demand for efficient and affordable therapies, speed to clinic/market is important, and biopharmaceutical companies push multiple drugs into development each year to ensure business sustainability (3,4,5,6).
Downstream purification process development for therapeutic MAbs is a critical step on their path to reach clinical trials and beyond (7,8,9). However, developing these processes under compressed timelines can be challenging because of a need to define many operating parameters and perform numerous empirical experiments that demand both time and resources. The most common MAb purification schemes use protein A affinity chromatography for capture, followed by one, two, or three chromatographic steps for polishing (10,11).
PRODUCT FOCUS: MONOCLONAL ANTIBODIES
PROCESS FOCUS: DOWNSTREAM PROCESSING
WHO SHOULD READ: PROCESS DEVELOPMENT AND ANALYTICAL
KEYWORDS: LIQUID HANDLING, SCALE-UP, CHROMATOGRAPHY, HOST-CELL PROTEINS, DNA ANALYSIS, UV-VIS SPECTROSCOPY
LEVEL: INTERMEDIATE
In addition, purification processes must be robust for viral clearance, with built-in viral inactivation and removal steps (12,13,14,15). In most cases, those process conditions need to be fine-tuned for each MAb and its process- or product-quality requirements.
Companies can increase process knowledge and reduce overall development time with increasingly efficient methods and tools for process development. Recently, high-throughput (HT) screening is gaining much attention with its potential for accelerating process development by enabling rapid acquisition of large datasets under many different operating conditions (16,17,18). HT screening has already been implemented successfully in the pharmaceutical industry to screen large numbers of small-molecule drug candidates (19,20,21,22,23) as well as in the biotechnology industry for discovery of affinity ligands for proteins from epitope libraries (24,25,26). However, use of HT technology in downstream process development is a more recent adaptation.
Kramarczyk was the first to describe the application of batch-binding chromatography in a 96-well filter plate format for development of protein purification steps using hydrophobic-interaction chromatography (HIC) and ion exchange (IEX) (27). Since then, tools for (or compatible with) HT operation have become prevalent in the bioprocess arena, and more continue to emerge. General process development operation needs that are successfully met by HT technology include liquid handling, buffer exchange, centrifugation, filtration, and vacuuming. Coupled with process development is in-process analytics, which has also developed numerous HT technologies: e.g., spectrophotometry, enzyme-linked immunosorbent assays (ELISAs), high-performance liquid chromatography (HPLC), capillary electrophoresis, and pH measurement.
Recently, preparative chromatography has been adapted to the HT format as well. This includes 96-well plate batch formats (28,29,30) involving wells as miniature columns and pipette tips filled with chromatography resins (e.g., from Atoll in Germany and Phynexus in San Jose, CA).
While defining our purification process, we sought to accelerate development by incorporating HT technology at key points. Figure 1 represents how we integrated HT technologies during process development. We used these technologies during early stages in development to scan a broad range of process parameters and increase our product and process knowledge before we initiated small-scale chromatography experiments. Examples of such experiments include buffer screening, resin screening, and identification of optimal wash and elution buffer conditions.Our resulting definition of an optimal operational space minimized the number of small-scale preparative chromatography experiments required to meet our ultimate product and process goals. Integration of HT technology into laboratory operations and purification development allowed rapid process development for an early phase molecule, which furthered our definition of a downstream operating space that could be applied to other biologics.
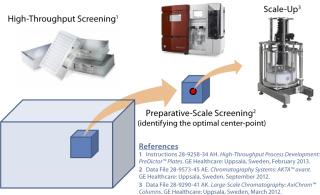
Figure 1: 1–3 ()
Materials and Methods
High-Throughput Liquid Handling and Chromatography: For HT liquid handling, we used a Tecan Freedom-evo system equipped with a high-precision Liquid-Handling (LiHa) arm, Robotic Manipulator (RoMa) system, and a Te-Stack module (Tecan Group, Ltd.). For method operations, we used Tecan freedom-evoware software. We executed HT chromatography experiments either in batch mode with resin plates packed using the Media Scout Resiquot device or column chromatography using prepacked 500-µL RoboColumn columns (Atoll GmbH).
High-Throughput Buffer Exchange and Size-Exclusion Chromatographic (SEC) Analysis: High-throughput buffer exchange for the MAb was performed using Zeba Spin 96-well desalting plates from Thermo Scientific (catalog #89808) according to the manufacturer’s protocol (Figure 2). Resin beds in the desalting plate were equilibrated with desired buffers and loaded with 80 µL of the MAb solution. Then the plate was centrifuged at 1,000g for two minutes before the flow-through was collected. We verified flow-through pH, ensuring complete buffer exchange, then incubated MAb in the plate at 19–25 °C for 24 hours. Size-exclusion ultraperformance liquid chromatography (UPLC-SEC) using a Waters’ Acquity H-Class Bio UPLC system measured the MAb’s stability in each buffer. We used an Acquity UPLC BEH200 SEC 1.7-µm column (4.6 × 150 mm) to perform that assay, in which the mobile phase consisted of 1× phosphate-buffered saline (PBS) at pH 6.8, run at a 0.4-mL/min flow rate.
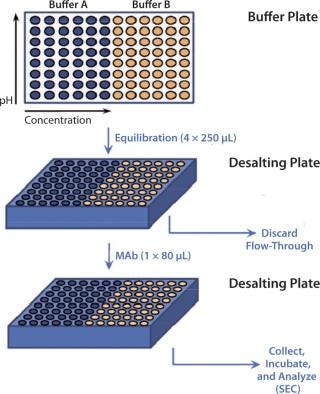
on:none;background-color: #f7f7f7;color:#000000;” >Figure 2: ()
The column remained at 25 °C throughout each run. We quantified monomer, low–molecular-weight species, and high–molecular-weight species using Empower software (Waters Corp.) and analyzed those results using JMP software (SAS Institute Inc.).
High-Throughput UV-Vis Spectroscopy: The HT UV-vis spectroscopy protein concentration assay used a DropSense96 polychromatic microplate reader (Trinean). We quantified the resulting UV-vis spectra using DropQuant software (Trinean). Unlike traditional clear-bottom 96-well UV-vis absorbance plates, DropSense plates required just 3 µL of samples. They eliminated the effects of meniscus-introduced changes in the pathlength that can be attributed to viscosity differences in samples.
High-Throughput ELISA — Chinese Hamster Ovary (CHO) Host-Cell Proteins: The Tecan liquid-handling system was used to perform a high-throughput ELISA for quantifying residual CHO host-cell proteins (CHO-HCP) with a CHO host-cell proteins third-generation kit (Cygnus Technologies catalog #F550) according to manufacturer’s protocol. An EnVision multilabel reader from PerkinElmer) measured absorbance at 450/650 nm using an Infinite M1000 PRO microplate reader (Tecan).
High-Throughput Analysis of Residual DNA: We used real-time quantitative PCR (RT-PCR) to quantify residual CHO DNA in the samples using a 2× TaqMan Universal PCR Master Mix kit (Life Technologies) and 7900 HT Real Time PCR system (Life Technologies) following the manufacturer’s instructions.
Preparative-Scale Chromatography: We used an ÄKTA Avant instrument (GE Healthcare Life Sciences) for preparative-scale, column-based chromatography experiments controlled by Unicorn 6.3 software (GE). All columns were packed in the laboratory according to the resin manufacturer’s recommendations.
Ahead in Part 2
Next month, part two will continue and conclude this article by reporting results and extensively discussing them. This includes nine figures and two tables of data.
About the Author
Author Details
Lead and corresponding authors Srinivas Chollangi (scientist) and Neil E. Jaffe (research investigator) contributed equally to this work; Hui Cai is a scientist I, Amanda Bell is an associate research scientist II, Krina Patel is a scientist II, Mark Fischl is an associate scientist I, Reb J. Russell is site director, Kuang-Chuan Cheng is associate director, and Michelle-Jue Wang is group leader, all in biologics development at Bristol-Myers Squibb, 519 Route 173 West, Bloomsbury, NJ 08804; 1-978-784-6248, fax 1-978-784-6639; [email protected], [email protected].
1.) Gottschalk, U 2006. The Renaissance of Protein Purification. BioPharm Int. 19:S8-S9.
2.) Reichert, JM. 2005. Monoclonal Antibody Successes in the Clinic. Nat. Biotechnol. 23:1073-1078.
3.) DiMasi, JA, RW Hansen, and HG Grabowski. 2003. The Price of Innovation: New Estimates of Drug Development Costs. J. Health Econ. 22:151-185.
4.) Datar, RV, T Cartwright, and CG Rosen. 1993. Process Economics of Animal Cell and Bacterial Fermentations: A Case Study Analysis of Tissue Plasminogen Activator. Biotechnol. 11:349-357.
5.) Sadana, A, and AM Beelaram. 1994. Efficiency and Economics of Bioseparation: Some Case Studies. Bioseparation 4:221-235.
6.) Clemento, A 1999. New and Integrated Approaches to Successful Accelerated Drug Development. Drug Info. J. 33:699-710.
7.) Werner, RG 2004. Economic Aspects of Commercial Manufacture of Biopharmaceuticals. J. Biotechnol. 113:171-182.
8.) Sommerfeld, S, and J Strube. 2005. Challenges in Biotechnology Production: Generic Processes and Process Optimization for Monoclonal Antibodies. Chem. Eng. Processing 44:231-236.
9.) Li, F. 2005. Current Therapeutic Antibody Production and Process Optimization. BioProcessing J. 4:1-8.
10.) Kelley, B, G Blank, and A Lee Gottshalk, U. 2009.Downstream Processing of Monoclonal Antibodies: Current Practices and Future OpportunitiesProcess Scale Purification of Antibodies, John Wiley and Sons, Hoboken:1-23.
11.) Vunnum, S, G Vedantham, and B Hubbard Gottshalk, U. 2009.Protein A-Based Affinity ChromatographyProcess Scale Purification of Antibodies, John Wiley and Sons, Hoboken:79-102.
12.) Zhou, J, and H Dehghani Langer, ES. 2007.Development of Viral Clearance Strategies for Large-Scale Monoclonal Antibody ProductionAdvances in Large Scale Biomanufacturing and Scale-Up Production, ASM Press/BioPlan Associates Inc, Rockville:1-28.
13.) Shi, L. 2004. Real Time Quantitative PCR As a Method to Evaluate Xenotropic Murine Leukemia Virus Removal During Pharmaceutical Protein Purification. Biotechnol. Bioeng. 87:884-896.
14.) Bray, J, and K Brattle. 2004.Monoclonal Antibody Production: Minimizing Virus Safety Issues, Volume 1, Plenum Publishers, New York.
15.) CPMP/BWP/269/95 2001.Note for Guidance on Virus Validation Studies, EMA, London.
16.) Coffman, JL, JF Kramarczyk, and BD Kelley. 2008. High-Throughput Screening of Chromatographic Separations: I, Method Development and Column Modeling. Biotechnol. Bioeng. 100:605-618.
17.) Kelley, BD. 2008. High-Throughput Screening of Chromatographic Separations: IV, Ion-Exchange. Biotechnol. Bioeng. 100:950-963.
18.) Kramarczyk, JF, BD Kelley, and JL Coffman. 2008. High-Throughput Screening of Chromatographic Separations: II, Hydrophobic Interaction. Biotechnol. Bioeng. 100:707-720.
19.) Gallop, MA. 1994. Applications of Combinatorial Technologies to Drug Discovery: 1, Background and Peptide Combinatorial Libraries. J. Med. Chem. 37:1233-1251.
20.) Gordon, EM. 1994. Applications of Combinatorial Technologies to Drug Discovery: 2, Combinatorial Organic Synthesis, Library Screening Strategies, and Future Directions. J. Med. Chem. 37:1385-1401.
21.) Van Hijfte, L, G Marciniak, and N. Froloff. 1999. Combinatorial Chemistry, Automation and Molecular Diversity: New Trends in the Pharmaceutical Industry. J. Chromatogr. B Biomed. Sci. Appl. 725:3-15.
22.) Janzen, WP 1996. High Throughput Screening As a Discovery Tool in the Pharmaceutical Industry. Lab. Robotics Automat. 8:261-265.
23.) Kenney, BA. 1998. The Application of High-Throughput Screening to Novel Lead Discovery. Progr. Drug Res. 51:246-269.
24.) Barrett, RW. 1992. Selective Enrichment and Characterization of High Affinity Ligands from Collections of Random Peptides on Filamentous Phage. Analyt. Biochem. 204:357-364.
25.) Oldenburg, KR. 1992. Peptide Ligands for a Sugar-Binding Protein Isolated from a Random Peptide Library. Proc. Natl. Acad. Sci. USA 89:5393-5397.
26.) Scott, JK, and GP. Smith. 1990. Searching for Peptide Ligands with an Epitope Library. Science 249:386-390.
27.) Kramarczyk, JF 2003.High-Throughput Screening of Chromatographic Resins and Excipients for Optimizing Selectivity, Tufts University, Medford.
28.) Lacki, KM 2012. High-Throughput Process Development of Chromatography Steps: Advantages and Limitations of Different Formats Used. Biotechnol. J. 7:1192-1202.
2
9.) Chatre, S, and NJ Titchener-Hooker. 2009. Microscale Methods for High-Throughput Chromatography Development in the Pharmaceutical Industry. J. Chem. Technol. Biotechnol. 84:927-940.
30.) Chatre, S, DG Bracewell, and NJ Titchener-Hooker. 2009. A Microscale Approach for Predicting the Performance of Chromatography Columns Used to Recover Therapeutic Polyclonal Antibodies. J. Chromatogr. A 1216:7806-7815.
You May Also Like