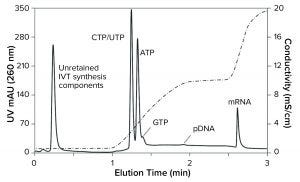
Figure 1: IVT analysis after four hours of incubation (CIMac PrimaS, see “IVT Characterization” box for conditions)
COVID-19 has focused a spotlight on the ability of mRNA technology to accelerate vaccine development and approval (1). That same technology can hasten development and approval of other therapeutic classes, including cancer immunotherapy, protein replacement, and gene therapy. Fulfilling those opportunities imposes significant challenges on process developers and manufacturers to improve existing processes. Scale-up to produce millions of doses (tens of kilograms) compounds those challenges. Furthermore, every step of the journey requires high-performance analytical methods, to ensure patient safety and maximize productivity.
A Complex Process
This complex process begins with a DNA plasmid (pDNA) produced in Escherichia coli and its subsequent purification. The process continues with in vitro transcription (IVT), followed by purification of the mRNA.
With the target DNA sequence defined and inserted into a plasmid, amplification is performed in E. coli to produce a pDNA template. Lysis is followed by precipitation with calcium chloride to reduce contamination by host-cell RNA. Plasmid purification by anion-exchange chromatography (AEC) and hydrophobic-interaction chromatography (HIC) follows to remove endotoxins, host-cell proteins, host-cell RNA, pDNA fragments, and aggregates (2). Then, supercoiled plasmid is linearized with an enzyme that requires subsequent removal.
Approaches to transcription of linearized plasmids are still evolving, but they essentially involve combining plasmids with the raw materials (nucleotides and enzymes) for synthesizing RNA (3). High efficiency of transcription is important because the more copies of mRNA that can be obtained from each pDNA molecule, the lower will be the ratio of product to contaminants in a completed IVT mixture. IVT production of mRNA for therapeutic use is complicated further by the need to stabilize the 5′ ends by adding a capping reagent either cotranscriptionally or posttranscriptionally.
Purification needs to remove the raw materials used for synthesis along with mRNA impurities such as aborted (short) transcripts, double-stranded RNA, and aggregates. Technologies for doing so also are evolving. As protein purification did in the past, the field of mRNA development is moving away from precipitation methods toward chromatographic methods that achieve higher recoveries and purities, more predictable scalability, and better reproducibility. Multiple options are available for both capture and polishing, including affinity, anion-exchange, hydrogen-bonding, hydrophobic-interaction, and reversed-phase chromatography methods (4–6).
The Analytical Bottleneck
The complexity of the entire processing chain from E. coli cell paste to purified mRNA is compounded by the less visible but equally important task of characterizing each step. It is fundamental to maximizing productivity and documenting the purity necessary to ensure patient safety. Characterization of IVT, especially as it applies to optimization, is particularly challenging because it must include interactions among many variables. The process begins with assessing the quality of reagents. Plasmids can carry proteins, endotoxins, and other residual contaminants. Similarly, enzymes can include stabilizing additives unsuitable for in vivo therapeutics, sometimes degraded to varying degrees (4–6). Nucleotides need to be provided in adequate quantities so that they do not limit transcription. The same is true of capping reagents. The ratio of enzymes to plasmids must be balanced carefully. Monitoring buffer conditions, time, and temperature also is essential. Some reagents are extremely expensive, so they need to be used under the most favorable conditions.
In the IVT process, the conditions outlined above require optimization or confirmation for every new mRNA construct. Common analytical methods used in IVT development provide information about one parameter at a time (e.g., agarose gel electrophoresis to confirm the presence of RNA species or sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to detect protein contaminants). Some of those methods are also time-consuming and lack automation, which extends development timelines. New analytical tools can potentially disrupt the way mRNA processes are developed and monitored.
Eliminating the Analytical Bottleneck
High-performance liquid chromatography (HPLC) has a long history of providing analytical data to support development and manufacture of biologics, and it has unique capabilities for mRNA development. Benefits of HPLC include rapid results, high resolution, and quantitative monitoring, with minimal sample volume requirements. Upstream processes such as IVT reactions, where the target molecule is produced and reagents are consumed, would benefit strongly from implementation of HPLC. AEC has been a foundation HPLC method for proteins and nucleic acids, but large mRNA molecules can be problematic because they do not elute from traditional AEC media at ambient temperature. New anion exchangers have begun to overcome that limitation.
Figure 1 illustrates ambient temperature fractionation of an mRNA IVT mixture at four hours from the start. The profile was produced with an ascending salt–pH gradient (for conditions, see the box at right). mRNA is well separated from other IVT components, which elute at earlier retention time. The profile also provides a snapshot of the levels of nucleotides and their proportions compared with those of pDNA and mRNA. The 100‑µL column format minimizes sample consumption, typically requiring sample volumes of only 25 µL (10–30 ng RNA). At the working flow rate of 20 column volumes (CV) per minute, a complete analytical cycle requires five to eight minutes. Profiles of this type provide the quantitative foundation necessary for evaluating complex reaction kinetics.
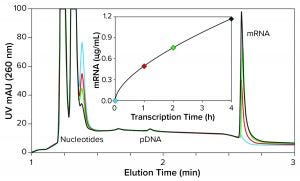
Figure 2: mRNA synthesis kinetics measured by anion-exchange HPLC analysis (CIMac PrimaS, see “IVT Characterization” box for conditions)
Figure 2 overlays IVT profiles from samples taken at 30 seconds, one hour, two hours, and four hours. Consumption of nucleotides is revealed in parallel with increasing concentration of mRNA. Analytical series of this type can be used to characterize quickly and efficiently the influence of each process variable desired.
PrimaS elution conditions also can be configured to highlight other contaminant subsets. As shown in Figures 1 and 2, plasmid DNA (pDNA) binds much more weakly than single-stranded mRNA (ssRNA). Double-stranded RNA (dsRNA) elutes later than pDNA but before mRNA (2–4). Weak binding also enables elimination of double-stranded species in a salt step, whereas elution of mRNA requires an increase of pH. PrimaS has the further ability to separate mRNA according to size in low-conductivity pH gradients. That enables discrimination of short transcripts and aggregates.
The role of AEC also extends to pDNA. Other anion exchangers are broadly used for initial monitoring of pDNA. Elution of pDNA from lDEAE, for example, takes place at ambient temperature. The supercoiled plasmid is clearly differentiated from open-circular and linear conformations and also from host RNA, aggregates, and proteins (6). AEC also provides an additional benefit for both pDNA and mRNA. Implementation of this new analytical method can provide improved process understanding, leading to faster, more robust and consistent manufacturing.
References
1 Wadhwa A, et al. Opportunities and Challenges in the Delivery of mRNA Based Vaccines. Pharmaceutics 12, 2020: 102; https://doi.org/10.3390/pharmaceutics12020102.
2 Černigoj U, Štrancar A. Scale-Up of Plasmid DNA Downstream Process Based on Chromatographic Monoliths. DNA Vaccines: Methods and Protocols. Sousa A, Ed. Humana: New York, NY, 2021; 167–192.
3 Hadas Y, et al. Optimizing Modified mRNA In Vitro Synthesis Protocol for Heart Gene Therapy. Mol. Ther. Met. Clin.Dev. 14, 2019: 300–305; https://doi.org:10.1016/j.omtm.2019.07.006.
4 Gagnon P, et al. Two New Capture Options for Purification of Large Messenger RNA. Cell and Gene Therapy Insights 6(7) 2020: 1035–1046; https://www.biaseparations.com/en/library/publications/1090/two-new-capture-options-for-improved-purification-of-large-mrna.
5 Gagnon P, et al. A New Runway for Purification of Messenger RNA. BioProcess Int. 18(10) 2020: 36–45; https://bioprocessintl.com/downstream-processing/separation-purification/a-new-runway-for-mrna-purification.
6 Gagnon P. Purification of Nucleic Acids: A Handbook for Purification of Plasmid DNA and mRNA. BIA Separations: Ajdovščina, Slovenia, 2020; https://www.biaseparations.com/en/products/monolithic-columns/books/226/purification-of-nucleic-acids-a-handbook-for-purification-of-plasmid-dna-and-mrna-for-gene-therapy-and-vaccines.
Tomas Kostelec is head of technical support; corresponding author Rok Sekirnik is head of process development, mRNA/pDNA, [email protected]; Anže Martinčič Celjar is a scientist; Kristina Šprinzar Nemec is project manager; Andreja Gramc Livk is head of process analytics development; Pete Gagnon is chief scientific officer; and Aleš Štrancar is managing director at BIA Separations, a Sartorius company.