Informatics Technologies in an Evolving R&D Landscape
Over the years, bioprocessing companies have leveraged a host of information technology (IT) to help them bring innovative new therapies to market. As the needs of a research and development (R&D) enterprise evolve, however, are such systems and applications a help or a hindrance? On one hand, the increasing sophistication of IT solutions — such as those designed to help users create advanced molecular models or track thousands of compounds through the discovery process — have enabled new levels of R&D innovation. On the other hand, at some point the law of diminishing returns kicks in as more resources and budgets become devoted to maintaining IT infrastructure (e.g., integrating information from different systems and databases and training people to use multiple applications).

Ultimately, organizations focused on developing new therapies need to be able to extract maximum value from their R&D data without overspending on informatics infrastructure that’s too cumbersome and inefficient. It’s at this point that R&D organizations can learn much from IT experiences in manufacturing and supply chain operations. In such environments, IT has been useful in life-cycle management solutions such as product life-cycle management (PLM) and enterprise resource planning (ERP) to drive greater efficiencies and collaboration, process automation, and improved information governance. Although optimizing the innovation life cycle may be similar in principle to PLM and ERP, the unique requirements of the R&D function — including highly scientific data — makes the optimization process very different in practice. Companies will need to decide on the best approach.
PRODUCT FOCUS: ALL BIOLOGICALS
PROCESS FOCUS: PRODUCTION, RESEARCH AND DEVELOPMENT
WHO SHOULD READ: INFORMATION TECHNOLOGY, EXECUTIVE MANAGEMENT, PROCESS ENGINEERS
KEYWORDS: PROCESS OPTIMIZATION, DATA MANAGMENT
LEVEL: INTERMEDIATE
Reducing the Complexity and Cost of Informatics
In today’s global and data-intensive R&D environment, IT infrastructure has become extraordinarily complex. Modern R&D enterprises now span multiple locations and areas of specialization, with numerous departments, disciplinary groups, and even individual scientists using sophisticated IT systems, applications, and other informatics tools. Consequently, when it comes time to streamline R&D from discovery to clinical trials and scale up, the resulting “patchwork quilt” that springs out from sharing data or deploying unified processes is an integration and maintenance nightmare. Valuable information can easily become lost inside “siloed” systems, equipment, and databases. Moreover, manual information management can be time-consuming, error prone, and expensive.
Enterprise-level informatics platforms facilitate data integration, process automation, and information sharing required for R&D activities such as compound management and modeling and simulation. It is vital that a platform be able to handle sources and types of R&D data. Some PLM and ERP systems offer structured row- and column-based data. Other systems include unstructured information such as scientific text and two-and three-dimensional models. Any solution should be able to capture and integrate data, run processes across it, and report results in a way that makes sense for multiple users from different departments.
Service-oriented architecture, cloud computing, and Web services have made those solutions possible. Web services can, for example, support plug-and-play integration of multiple data types and formats without requiring customized (and expensive) IT intervention. Moreover, processes such as analysis and reporting steps can be broken into “parts” that can be put together in different ways depending on a person’s specific requirements. Because those parts can function independently from their source system or application, they can be used to create automated workflows that cross departmental or disciplinary boundaries.
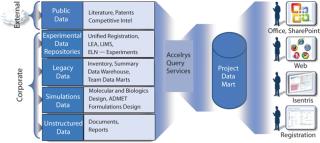
Figure 1: ()
A holistic informatics infrastructure that encompasses the needs of an entire R&D enterprise offers time, cost, and efficiency benefits. Regardless of where or how information is generated, it can be used by numerous contributors across the innovation life cycle, enhancing collaboration and speeding cycle times. Toxicologists can make their history of assay results available to chemists investigating new lead compounds, for example. Chemists can work more closely with sourcing experts to ensure that compounds in development are viable candidates for large-scale production.
In addition, processes that were previously disjointed (because critical data were locked within isolated databases and proprietary systems) can be streamlined and automated. Because information access is available, “nonexperts” also can take better advantage of sophisticated R&D knowledge (e.g., predictive modeling and analytics) that they may have been unable to use before without expert help. Moreover, R&D technologies leveraged throughout an organization are much easier and less costly for IT to manage and maintain because manual, hand-coded integration is no longer required. The result is that more resources can be devoted to high-value information analysis and less on the “care and feeding” of IT infrastructure.
Enterprise Informatics in Action
Accelrys has developed an enterprise R&D architecture built on the open services-based Pipeline Pilot informatics platform to address challenges of infrastructure complexity, data overload, and disjointed processes. Rather than trapping information in “silos,” the platform enables integration of diverse systems, applications, and data sources. It supports a collection of tools for manipulating and analyzing complex information, including advanced chemistry, biology, and materials science. The end-to-end, enterprise-level approach
enables scientists, engineers, and other R&D project stakeholders to easily access, analyze, report, and share data across departments and disciplines
frees IT from manually supporting varied requirements of multiple information consumers
allows an organization to extract maximum value from the many sources of innovation knowledge, both internal (e.g., data from experiments, modeling, and simulation) and external (e.g., publicly available databases or published literature).
The Cheminformatics suite is a key component of the enterprise R&D architecture and a first step toward innovation life-cycle management. It provides end users with a unified solution for registering, searching, mining, and analyzing information about chemical structures and reactions, chemically modified sequences, biological entities and associated data such as inventory, assay, and experimental results. Users have real-time access to pertinent chemical and biological information and an integrated environment for data analysis and reporting, with reduced IT overhead and management of multiple vendor relationships. The solution can serve as a solid infrastructure foundation that companies can build on over time. A company’s IT staff can take a brick-by-brick approach to addressing informatics demands, plugging new tools into the framework as needed. The enterprise scale of the platform lowers the complexity of IT architecture to ease overall total cost of ownership, planning, and investment decisions.
Example: One leading global pharmaceutical research company has benefited from more holistic and integrated informatics (1). The organization wanted to unify data analysis and collaboration activities throughout its medicinal chemistry design cycle and used Accelrys technology to power an underlying informatics foundation. The goal was to decrease the time required to capture, manage, integrate, and analyze data related to both real compounds that had already been synthesized, as well as information connected to design hypotheses associated with virtual compounds. The organization also sought to improve collaboration and decision-making through automated project workflows and communication spanning hypotheses creation and compound design, on through to synthesis, testing, analysis and further design.
Accelrys’ Pipeline Pilot platform was used to automate the extraction, transformation, and loading of data from multiple sources into project-specific data marts that can then be leveraged for “one stop shopping” analysis and decision-making. The types of data were varied and included primary asset data, in vitro absorption, distribution, metabolism, excretion, and toxicity (ADMET) data, precalculated molecular property data, in vivo ADME endpoint data, compound registration information, and in silico model calculations for both real and virtual compounds. In addition to data integration, automated protocols were also used to facilitate analysis, register new molecules, and determine compound uniqueness.
In this particular case, the underlying platform interoperated with several diverse third-party and legacy systems that project teams used throughout the discovery process for analysis and reporting. However, the evolution of the enterprise R&D architecture, including the release of the latest version of the Cheminformatics suite, enables users to take advantage of a host of built-in analysis, modeling, and reporting tools. This doesn’t preclude IT from also plugging in other applications and systems, but it does help the company move closer to a single vendor and R&D architecture that is easy to maintain and manage.
Facilitating “Open Innovation”
What happens when the innovation life cycle extends beyond a single organization? In this era of globalization, R&D stakeholders are more likely to be communicating across time zones and continents than across a conference table or laboratory bench. Increasingly, project participants no longer all work under the same corporate umbrella. Biopharmaceutical development can involve a host of contributors, including big brand names, smaller specialized companies, academic institutions, and government entities.
This is the realization of a concept known as open innovation. Instead of doing everything in-house, organizations are looking for ways to expand the scope, accelerate the speed, and increase the quality of their efforts by collaborating more extensively with external partners and sharing knowledge and expertise for their mutual benefit. For example, scientists in far-flung locations, niche experts, or contract research providers who can more inexpensively run assays or make compounds may all contribute to a single project. To practice efficient and cost-effective open innovation, companies must consider how best to leverage their informatics infrastructure to accommodate project participants both inside and outside their organizational boundaries.
Here again, the principles of life-cycle management apply. Software solutions for PLM, ERP, and supply-chain management (SCM) are all about driving efficiencies and bringing governance to complex networks involving numerous moving parts and participants. There are some key practical differences between these solutions and the needs of R&D, however. Innovation is not like a just-in-time supply chain. It depends on free-form interactions, includes processes that are constantly evolving, and resists too much structure.
That’s why smaller organizations are often hotbeds of creativity and innovation. It’s easy for stakeholders to kick ideas around at a company lunch table or visit a colleague across the hall. But when the project manager is in Boston, the lead chemist works for contract research organization in Beijing, and manufacturing is set up in India, the level of fluid communication and open dialogue required for open innovation is not so simple. Today’s R&D organizations need, therefore, to be able to recreate the open, collaborative atmosphere that existed when project participants were in the same building, but on a scale that embraces the breadth and complexity of a global market.
The advent of cloud computing is facilitating global collaboration. By enabling organizations to run applications, platforms, even underlying IT infrastructure as a service accessed through a Web browser, this computing paradigm is fundamentally changing how data are stored, shared, and managed. Many of us use cloud computing on a regular basis without even thinking about it; e.g., when we check a Gmail account, or share photos on a site such as Flickr. Instead of installing another e-mail application on our hard drive, we use Google’s; instead of copying photos to a disk and mailing them to friends and relatives, we send a Web link. Apply this same concept to open innovation, and the cloud becomes an ideal forum for project stakeholders to interact and share ideas regardless of where they are located, the organization they are with, or how much data are involved.
For example, suppose a pharmaceutical company is working with a contract research organization on a drug discovery project. It could install its legacy IT systems at the outsourcer’s site to exchange and analyze data. But this is costly and highly inefficient because systems must be maintained both within the organization’s internal IT infrastructure as well as at the CRO site. The redundancies multiply as more departments, locations, and partners that are included. By contrast, cloud computing is simple to access from numerous global sites. It’s also infinitely scalable, capable of accommodating more data and more participants as a project’s needs expand. The end result is less infrastructure and more analysis.
Cloud-based collaboration also can support a more open and vibrant CRO market. When it’s easier to pass information back and forth between partners, organizations can quickly bring in CRO expertise needed for even a single screen or test. This is especially compelling for smaller biotech companies that can leverage the cloud to manage projects and optimize intellectual property without having to increase internal headcount for basic research and IT services. Thus, small companies can compete more aggressively with bigger players.
Cloud Collaboration in Action
Cloud-based technologies are now making open innovation among numerous partners a reality with minimal IT overhead and effort. A
n excellent example is the use of the SCYNEXIS Hit Explorer Operating System (HEOS) software in conjunction with Accelrys’ Cheminformatics suite to empower a variety of diverse contributors and sites working on the Drugs for Neglected Diseases initiative (DNDi) (2). The HEOS system, supported by Enterprise R&D architecture, is a cloud-based, software-as-a-service (SaaS) workspace that supports real-time data sharing among multiple organizations engaged in collaborative drug discovery.
In the case of DNDi, the technology has enabled some breakthroughs in the research and development of better treatments for underfunded yet devastating diseases such as malaria, Chagas, schistosomiasis, and African trypanosomiasis (sleeping sickness) that affect millions of people in the developing world. Using the HEOS and Accelrys solutions, researchers from Brazil, the Ivory Coast, the Philippines, South Africa, Zimbabwe, and many other countries have been able to come together in a vibrant virtual community to contribute their insights and expertise to this important project. Regardless of their physical location, disciplinary focus, experience, or cultural background, participants can exchange and analyze complex biological and chemical data, assay results, computation and visual simulations, and more. The group as a whole can investigate and test a wider diversity of compounds and therapies, which can greatly speed R&D efforts.
One initiative spearheaded by DNDi against an endemic disease ultimately included multiple levels of collaboration between volunteers from large pharma and biotech companies, contract research organizations, and academia. The biotech company that provided the initial compounds managed the initiative, tracked in vivo studies performed by a US university, and brought on a CRO that would confirm the most promising leads through high-throughput testing. As the project continued, several ADME, safety, and pharmacokinetic teams got involved, and additional peer organizations were consulted. The end result was a compound that today shows great promise in addressing a disease that hasn’t seen a new treatment in decades.
The case of the neglected diseases technology deployment demonstrates that complex data can be stored securely and shared globally on a thin client. However, the real takeaway is that improved global collaboration in cloud computing is powering the kind of open innovation that represents the next evolution of R&D. In the years ahead, the scope and scale of R&D enterprises are only going to get broader.
The organizations best equipped to succeed in this complex, global, and multilayered landscape will be those that, like their counterparts in manufacturing, start laying the groundwork for a more holistic, streamlined, flexible, and open informatics infrastructure today. Cloud computing, service-oriented architecture, and Web services can facilitate manipulation of highly scientific information and innovative life-cycle management that streamlines R&D. An ability to focus on high-value analysis and minimize resources that go into data integration and systems maintenance will ultimately enable faster and cheaper time-to-innovation. And an ability to bring all valued contributors to the table, whether across the hall or on the other side of the world, will drive more extraordinary and groundbreaking discoveries.
About the Author
Author Details
Frank Brown, PhD, is executive director at Merck and former executive vice president and CSO and Accelrys. Corresponding author Matt Hahn, PhD, is senior vice president and CTO at Accelrys; [email protected].
REFERENCES
1.) Brodney, MD. 2009. Project-Focused Activity and Knowledge Tracker: A Unified Data Analysis, Collaboration, and Workflow Tool for Medicinal Chemistry Project Teams. J. Chem. Inf. Model. 49:2639-2649.
2.) Bost, F, RT Jacobs, and P. Kowalczyk. 2010. Informatics for Neglected Diseases Collaborations. Drug Discovery Devel. 13:286-296.
You May Also Like