- Sponsored Content
- Expression Platforms
Quantitative Synthetic Biology for Biologics ProductionQuantitative Synthetic Biology for Biologics Production
Sponsored Content
Biologics accounted for more new drug approvals than did small molecules for the first time in 2022, marking a significant shift in the pharmaceutical industry (1). Large-molecule pipelines are also moving from standard monoclonal antibodies (MAbs) to more complex and difficult-to-express molecules, which intensifies pressure on the industry to meet biomanufacturing demands. There is a pressing need for innovative Chinese hamster ovary (CHO)–based bioproduction systems to keep pace with this evolving landscape.
While multiple areas of cell-line development (CLD) have improved over the years, advances in expression vector design have lagged behind other technologies. Most expression vectors still rely on a “one-size-fits-all” approach across molecules in which the coding sequences (CDSs) of different therapeutic proteins are inserted into fixed plasmids made up of legacy genetic parts. That method often results in suboptimal expression, increased manufacturing costs, and delays in clinical development for both MAbs and other, more complex modalities.
Asimov’s CHO Edge system builds on the current state of the art for CLD by integrating expanded genetic tools with data-driven models. The system includes a glutamine synthetase (GS) knock-out CHO host — and a Fut8/GS double knock-out also is available for afucosylated antibody production — with a proprietary hyperactive transposase for genomic integration, a library of more than 2,500 characterized genetic elements, and Kernel computer-aided design software for vector design and simulation.
The integrated system routinely achieves titers of 5–10 g/L across modalities in a four-month CLD timeline (Figure 1). The entire system can be licensed and also is offered as a CLD service with a disruptive commercial structure. The cost of a campaign is linked directly to research cell bank (RCB) performance: If a generated RCB expresses a MAb at <4 g/L, then the CLD campaign and all commercial-use rights will be free of charge.
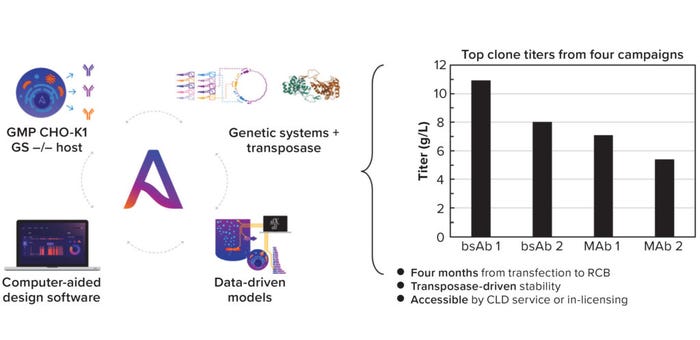
Figure 1: (left) Asimov’s CHO Edge system comprises a good manufacturing practice (GMP) CHO-K1 GS knock-out host, a library of genetic parts, and a hyperactive transposase, with data-driven models and a unifying software interface for design and plasmid request; (right) top clone titers for four different molecule CLD campaigns.
Expression Vector Design
Genetic Parts: An expression vector’s genetic parts significantly impact a molecule’s expression. For example, MAbs often are expressed using identical promoters for both their heavy and light chains, which can limit expression titers (2, 3). Asimov has developed a growing library of more than 2,500 characterized genetic parts for use in a proprietary CHO host cell line. Those parts span many different functions, including constitutive promoters, untranslated regions (UTRs), epigenetic insulators, internal ribosome entry sites, signal peptides, polyadenylation signals, and small-molecule inducible systems (Figure 2a). The breadth of behaviors we have quantitatively characterized enables precise control over biologics expression.
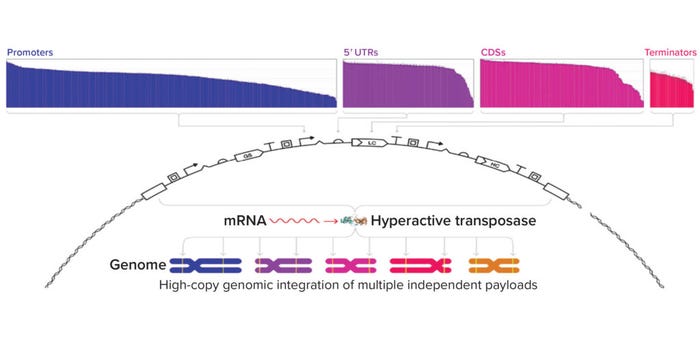
Figure 2a: Expression vectors are customizable with a library of more than 2,500 characterized genetic parts spanning diverse cell functions. Vectors are integrated into the genome through a proprietary hyperactive transposase.
Even with a large library of genetic parts, however, manual vector design remains difficult because of the number of possible selections and vector arrangements. We developed a computational genetic simulator that models biophysical phenomena arising from an expression vector, such as transcription, translation, and regulation (Figure 2b). Note that expression vectors can be simulated only if they are made up of characterized genetic parts from our database. Our simulation capabilities will continue to advance as additional experimental data and biophysical phenomena (e.g., epigenetics) are incorporated into the model.
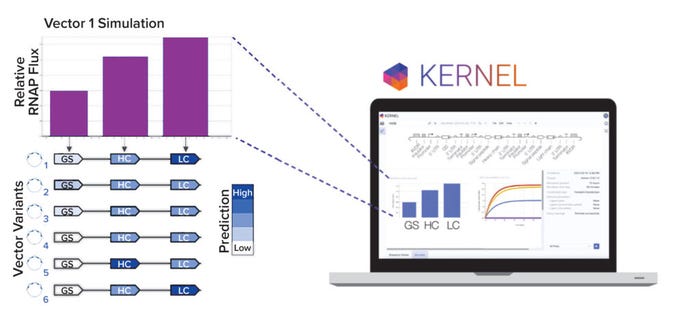
Figure 2b: Kernel computer-aided design software is used to model vector variants computationally. A panel of six variants was designed to explore heavy-chain/light-chain ratios and selection stringency.
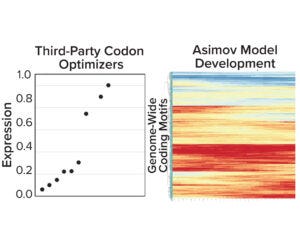
Figure 3a: (left) Gene expression results from five third-party codon optimizers; (right) Asimov’s holistic CDS optimizer accounts for positional effects within the CDS and other biophysical phenomena.
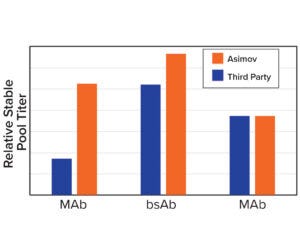
Figure 3b: Asimov’s CDS optimizer results in higher titers compared with leading third-party algorithms.
Coding-Sequence and Signal-Peptide Optimization: To gain a deeper understanding about the importance of codon optimization, we benchmarked five third-party codon optimizers for the same protein and documented a 20-fold difference in expression across CDS variants (Figure 3a, left). Those data motivated us to develop a holistic CDS optimization algorithm that goes beyond traditional codon-frequency–based methods. Our algorithm incorporates sequence features based on mechanistic models of transcription and translation, CDS positional effects, secondary structure, and other biophysical parameters (Figure 3a, right). CDS optimization led to significant improvements in expression across multiple molecules compared with a leading third-party codon optimizer (Figure 3b).
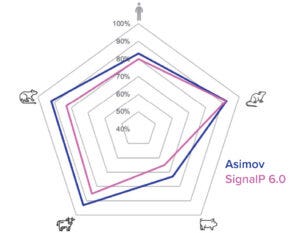
Figure 3c: New signal-peptide model has higher accuracy than the SignalP 6.0 algorithm.
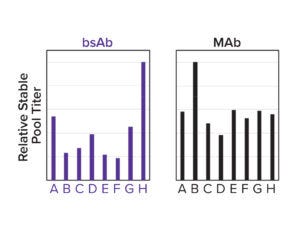
Figure 3d: A panel of designed signal peptides shows differential titers across two molecules.
To both ensure high cleavage efficiency and increase expression further, a signal-peptide prediction tool was developed based on machine-learning technology. The algorithm integrates a deep-learning architecture with a large protein language model that outperforms the industry-leading SignalP 6.0 algorithm in prediction accuracy across mammalian species (Figure 3c) (4). We used this model to design a panel of signal peptides that were predicted to exhibit precise and efficient cleavage. Pairing those signal peptides with two different molecules, we generated stable pools and observed a better than fivefold titer difference between the least and most optimal pairings (Figure 3d). The signal peptides can be selected with the Kernel software to maximize expression on a protein-specific basis.
Cell-Line Development Data
By leveraging the above tools, we designed a set of expression vectors to explore the impact of genetic-part selections on the expression of different two-chain molecules. Those vector variants modulate the ratios of heavy to light chains and stringency of selection through GS expression.
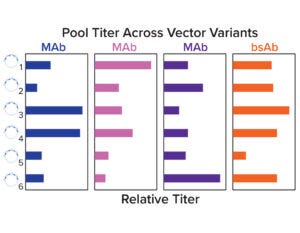
Figure 4a: Relative stable pool titers across a panel of six vector variants.
Stable pools were generated for a set of four molecules across six vector variants by cotransfecting each plasmid with chemically modified mRNA encoding a hyperactive transposase. Pool titers show that the optimal vector configuration was molecule dependent, and no single variant had high performance across all molecules. Both selection stringency and heavy-chain/light-chain ratios were deemed to be important for expression (Figure 4a).
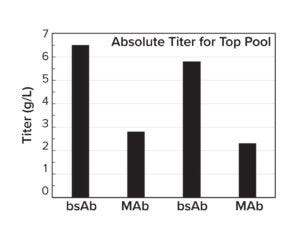
Figure 4b: Stable pool titers in a 14-day Ambr 15 fedbatch process.
All top stable pools achieved 2–7 g/L in a 14-day, fed-batch, Ambr 15 microbioreactor run (Figure 4b). That vector dependence suggests that the conventional, one-size-fits-all approach has inherent limitations and highlights the importance of customization to meet the unique requirements of each therapeutic protein candidate. Results generated from internal CLD campaigns can be incorporated as training data into the simulator to guide future CLD projects.
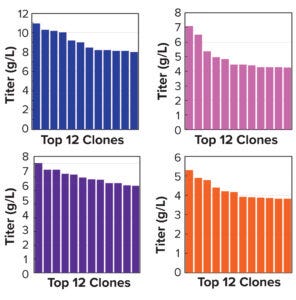
Figure 5a: Clonal titers in a 14-day Ambr 15 process.
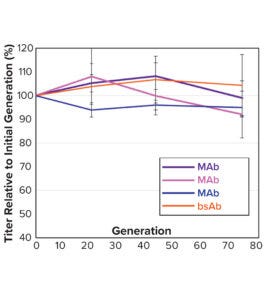
Figure 5b: Average relative titer of five to seven clones per molecule for four separate CLD campaigns (standard deviation shown in the error bars); all titers were measured with an Octet analysis system.
Note that vector design space can be explored efficiently without compromising CLD timelines. All stable pools are generated in parallel, with only the top performers advanced to single-cell cloning. Selected clones from the above pools were evaluated using a 14-day Ambr 15 fed-batch process, and they all achieved expression titers of 5–10 g/L (Figure 5a). We used protein-A purified material to confirm high product quality, including monomeric content, charge variance, and glycan structure (data not shown). For all four campaigns, clonal distributions were relatively homogenous, and top clones were not statistical outliers. Genomic sequencing of those clones showed that transposase-mediated integration resulted in high-copy integration of multiple independent payloads. In addition, clonal titers should increase further with process and media optimization. Finally, the top clones maintained >80% titer stability over 75 generations, which suggests that stability studies could be removed from the critical path of development (Figure 5b).
Advancing Biologics Production
Asimov’s CHO Edge system integrates a GS knock-out host cell line with a hyperactive transposase, a diverse and growing genetic-parts library, and data-driven computational models to advance biologics production. By exploring expression vector design space, the system can achieve clonal protein expression titers exceeding 10 g/L. Planned future capabilities include the extension of models to optimize cell culture media and bioreactor conditions, with an ultimate goal of developing an end-to-end toolbox for CLD and process development guided by in silico technology.
References
1 Senior M. Fresh from the Biotech Pipeline: Fewer Approvals, but Biologics Gain Share. Nature News 9 January 2023; https://www.nature.com/articles/s41587-022-01630-6.
2 Schlatter S, et al. On the Optimal Ratio of Heavy to Light Chain Genes for Efficient Recombinant Antibody Production by CHO Cells. Biotechnol. Prog. 21(1) 2005: 122–133; https://pubmed.ncbi.nlm.nih.gov/15903249.
3 Zhang J-H, et al. Strategies and Considerations for Improving Recombinant Antibody Production and Quality in Chinese Hamster Ovary Cells. Front. Bioeng. Biotechnol. 4 March 2022; https://www.frontiersin.org/articles/10.3389/fbioe.2022.856049/full.
4 Teufel F, et al. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 3 January 2022; https://www.nature.com/articles/s41587-021-01156-3.
Haewon Chung, Brianna Jayanthi, Alina Ferdman, Georgian Tutuianu, Jeremy J. Gam, Kevin D. Smith, Niko McCarty, Raja Srinivas, and corresponding author Alec A.K. Nielsen ([email protected]) are with Asimov, Inc., 201 Brookline Avenue, Suite 1201, Boston, MA 02215; 1-617-849-9299; [email protected]; https://www.asimov.com. Formerly with Asimov, Dinghai Zheng is now at Sanofi.
You May Also Like




.jpg?width=700&auto=webp&quality=80&disable=upscale)

