Alltrna recently built a platform designed to turn the sophisticated biology of tRNA into programmable medicine. This technology aims to enable scientists to use the therapeutic properties of tRNA to accelerate the development of genetic medicines for millions of people with rare diseases that are caused by the same underlying mutation.
William Kiesman, chief technology officer of Alltrna, spoke with BioProcess Insider about the role tRNA can have on human health and disease beyond serving as amino-acid–shuttling intermediaries in protein translation.
BioProcess Insider: Can you describe what tRNAs are?
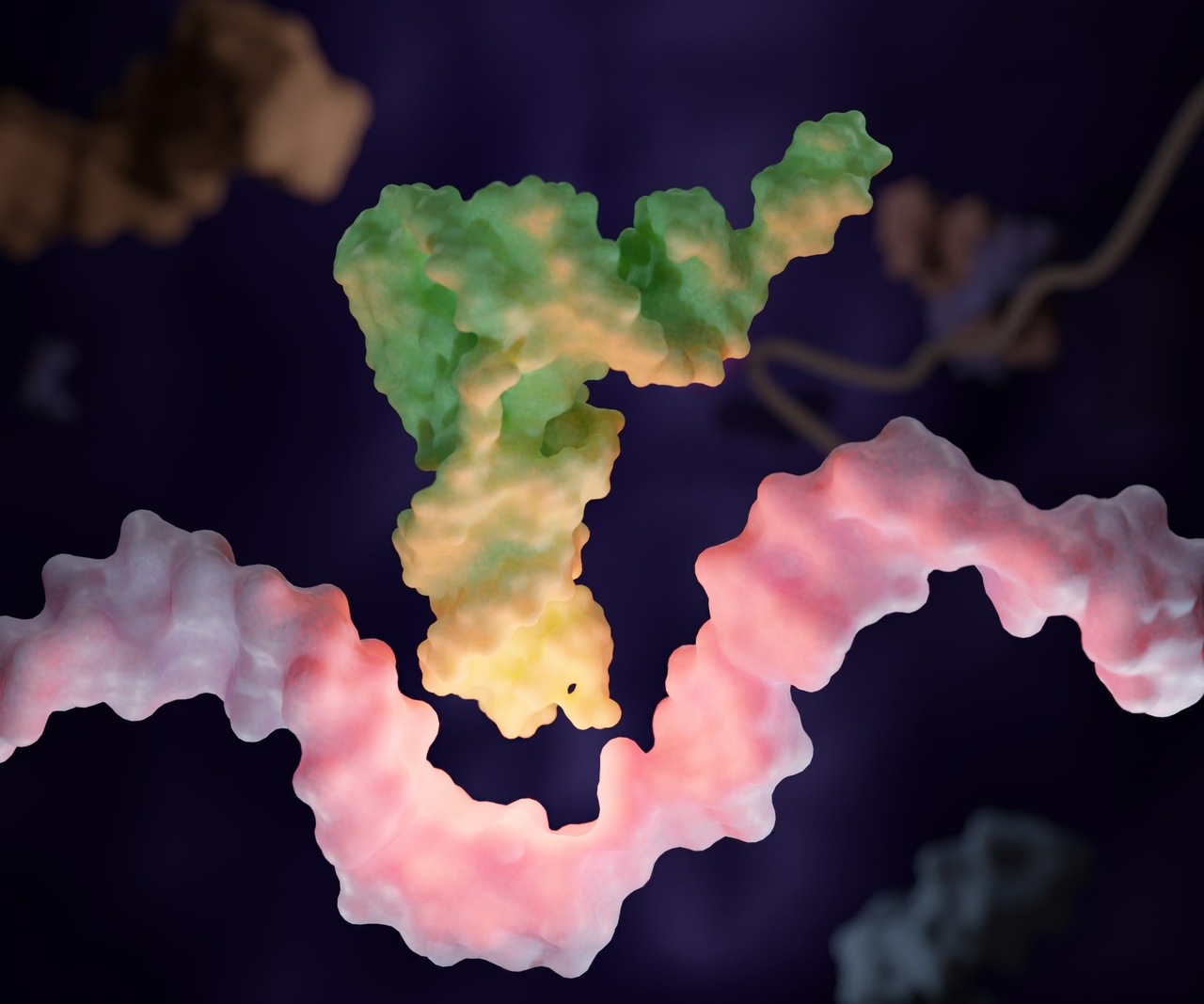
William Kiesman (WK): Fundamentally, tRNAs are short, highly structured RNA molecules that serve as a central component of the protein translation machinery in a cell. Each tRNA is conjugated to an amino acid, the identity of which is determined by the tRNA’s sequence.
When a gene is expressed, a messenger RNA (mRNA) copy of the gene carries a code based on nucleotide triplets (codons) that dictates the amino acid sequence of the protein to be produced. Working with ribosomes, the tRNA uses its anticodon sequence to read the mRNA codons, and when they complement, the tRNA transfers its amino acid to the growing polypeptide chain.
This process is repeated until the complex reaches a special codon that has no corresponding tRNA – a natural termination or stop codon – at which point, protein translation stops, and a fully formed, functional protein is released.
BPI: In layman terms, how are tRNAs manufactured?
WK: tRNA manufacturing is a lot like assembling a string of beads one bead at a time. In this case, however, each of those beads – the nucleotides – is modified to ensure that the chain can only be constructed in one particular way. In trying to make a linear string of nucleotides, we want to make sure they don’t branch out in other directions.
The initial nucleotide is attached to a solid support structure that simplifies each round of nucleotide addition and purification. The attached nucleotide is then adjusted to ensure that when it is mixed with a solution of the next nucleotide in the sequence, they join the right way to form what is known as a dinucleotide. Once this has happened, the excess free nucleotide in solution is washed away to prepare the bead-bound dinucleotide for the next nucleotide and so on.
The process is repeated with each subsequent nucleotide in a specific order until the tRNA reaches the target length. It is then removed from its solid support. We then purify the full-length tRNAs from shorter incomplete fragments and other contaminants.
BPI: What can tRNAs be used for?
WK: A primary job of tRNAs and their attached amino acids is to extend the polypeptide chain by decoding mRNA codons, only stopping when the process reaches a natural termination codon. Sometimes, however, a mutation occurs that changes an amino-acid–coding codon into a premature termination codon (PTC). When this happens, protein translation stops early, producing either no protein or a truncated and possibly toxic protein, which can have deleterious effects in the body. We call the diseases that arise from these mutations stop codon diseases, which affect more than 30 million around the world.
Alltrna is designing unique tRNA molecules that can read PTCs and insert the amino acid that would have been present if not for the mutation. This approach aims to rescue protein translation and thereby reverse disease. Our approach is unique because, rather than treat disease mutation-by-mutation one gene at a time, our engineered tRNAs are gene-agnostic, focusing instead on the PTC mutation, which might be shared between hundreds of Stop Codon Diseases.
Most stop codon diseases are rare or ultra-rare, making drug development less financially feasible due to small patient populations. Because of that, the ability to develop a single treatment against multiple diseases will be transformative. Already, we have shown that a single, purpose-built tRNA can rescue protein expression in 25 different disease models across 14 different genes regardless of where the mutation occurs in the gene.
Beyond the stop codon diseases, we see potential for tRNA-based medicines against other types of mutations or to influence what proteins are expressed by adjusting the pool of available tRNAs in a cell. And our understanding of the roles of tRNA fragments in biology and disease is constantly expanding, offering future therapeutic avenues for us to explore.
BPI: What are the advantages of using tRNAs?
WK: One that was especially helpful to us is tRNA’s incredible diversity across all life forms. Although nature really only requires 61 different tRNA genes to address the 61 amino-acid-coding codons, many organisms have hundreds of tRNA genes. Humans, for example, produce more than 270 different types of tRNA molecules from about 450 unique and duplicated tRNA genes, while zebrafish, for example, have more than 12,000 tRNA genes.
That diversity means there is a huge amount of flexibility in tRNA sequence without perturbing their function. It empowers Alltrna to explore more of the sequence space to design therapeutic molecules optimized not only for biological activity, but also for tolerability, stability, manufacturability, and a host of other characteristics required in drug design. Instrumental to understanding, leveraging, and expanding this diversity is our expertise in machine learning and computational chemistry.
Compared with other RNA-based medicines, molecular size is also an advantage in designing therapeutic tRNAs. Gene and mRNA-based therapies may contain thousands of nucleotides; for example, the mini-dystrophin gene is 6,000 to 8,000 nucleotides. This large size can complicate formulation with delivery vehicles like lipid nanoparticles and limit the therapeutic payload – the amount of medicine that can be delivered in any single particle. By comparison, tRNAs are quite small at 70 to 80 nucleotides, allowing us to package more molecules into each LNP and thereby influence the dose levels and dosing frequencies.
BPI: Are there any challenges associated with tRNAs?
WK: The chemical synthesis of longer RNAs, like tRNAs has historically been a challenge. The chemo-enzymatic and in vitro transcription (IVT) methods used to produce the much larger mRNAs didn’t work as well for tRNAs because the enzymes used to couple the nucleotides are not well suited to the coupling of chemically modified nucleotides that are a mainstay of the tRNA composition. Thus, we put a lot of effort into improving RNA synthesis technologies to ensure efficient, reproducible, and scalable production.
Beyond size, another challenge was ensuring we could optimize and produce the myriad chemical modifications that, like the nucleotide sequence, significantly influence tRNA structure and activity. Being able to identify these modifications and their positions within the tRNA molecules, and then use machine learning tools to tease out their biological importance was a critical early step in our efforts to engineer tRNA medicines.
BPI: Where does the manufacturing of tRNAs take place for Alltrna?
WK: We have built an end-to-end development scale capability internally that can supply drug substance and drug product for research studies and have put in place a network of CMO partners that can manufacture and supply toxicology and cGMP tRNA drug substances and drug products. In addition, we have laid the groundwork for a host of analytical techniques that we use to characterize the manufacturing processes and release the drug substance and drug product. While we supply the initial development pipeline, we are also embarking on novel process development for the synthesis, purification, and drug product unit operations to ensure that we can manufacture high quality products in the most sustainable ways possible.
BPI: What is Alltrna’s goal with tRNA production?
WK: We are initially looking to transform the therapeutic options for millions of underserved or unserved patients living with stop codon diseases, and from there, to expand into myriad other conditions that could benefit from engineered tRNAs or tRNA fragments.
About the Authors
You May Also Like








