Microbial Expression and Purification: One Company’s Historical PerspectiveMicrobial Expression and Purification: One Company’s Historical Perspective
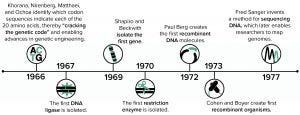
Figure 1: Early advances in DNA understanding were foundational for protein overexpression.
Since the dawn of the recombinant DNA era in the 1970s, New England Biolabs (NEB) has been integrally involved in expressing and purifying proteins, both for its own research interests and for biomanufacturing processes. In 1978, the company began screening microorganisms for restriction enzymes. Our scientists remember the challenges met in purifying limited amounts of restriction enzymes and other proteins from native organisms isolated from the environment. The efforts of those scientists to clone, overexpress, and purify restriction enzymes from recombinant systems helped to advance the field of molecular biology. Many of their original methods have endured to be applied by other scientists in studying the structure and function of individual proteins. Now, NEB scientists are striving to develop rapid, simplified methods for recombinant protein expression and purification that rely on engineered protein expression hosts and optimized cell-free systems.
The period from 1966 to 1977 brought a series of remarkable breakthroughs in biological science (Figure 1). During that time, the genetic code was interpreted correctly, the first gene was isolated, and enzymes that both cut DNA at specific sequences (restriction enzymes) and that paste DNA pieces together (DNA ligases) were discovered. Those discoveries ultimately enabled the first gene cloning and creation of the first genetically modified microorganisms. In 1977, DNA sequencing technologies advanced beyond the laborious extension of just a few bases at a time to provide scientists with the ability to unlock the genetic information encoded in a given segment of DNA. The remarkable scientific advances in that decade made protein overexpression and purification possible, forever changing the course of biological and medical research while enabling the biotechnology industry to emerge.
NEB was founded in 1974 with a goal of providing researchers with purified restriction enzymes, DNA ligases, and other tools needed to clone and express genes. Restriction enzymes were the cornerstone of our early product offerings. At that time, our scientists purified them from bacteria isolated from the environment. That presented many challenges for commercial-scale production. For example, native restriction enzymes generally are not expressed abundantly and must be purified free from many other nucleases that wild-type organisms produce. Additionally, difficulties often arose with large-scale culturing of obscure microorganisms.
To meet a steadily growing demand for such molecular tools — and to lower costs for customers — our company turned to recombinant DNA technology to clone and express enzymes in the familiar laboratory bacterium, Escherichia coli. NEB produced some of the first recombinant enzymes available for commercial sale, which began the company’s long experience with recombinant protein expression. Since those early days, that technique has been integral to our success and to biotechnology in general.
Over the past 40 years, we have worked continuously to invent and adopt new expression methodologies that could improve production of recombinant proteins — commercializing more than 550 recombinant enzymes to date. Below, we highlight some major innovations in protein expression that have driven our company’s journey, taking both historical view and an eye toward the future.
Early Recombinant Protein Expression
The first recombinant enzymes from E. coli were offered for sale in 1980: DNA polymerase (Pol I) first cloned by Bill Kelly in Noreen Murray’s laboratory at Edinburgh University, and T4 DNA ligase, which had been cloned by Geoff Wilson in the same laboratory several years earlier. Dedicated research on protein expression also began at NEB that year, including efforts to create a vaccine against malaria using recombinant surface antigens from parasites. The company adopted new cloning and expression methodologies for use with restriction enzymes to increase yields, improve purity, and facilitate characterization of restriction-enzyme structure and function.
Early work involved establishing methods and tools to enable restriction enzyme cloning in E. coli, which already had become the standard for cloning and expression and remains so today for proteins that do not require complex posttranslational modifications (1). To clone foreign restriction-modification systems in E. coli and overproduce individual restriction enzymes, it was necessary to characterize and eliminate the organism’s native methyl-dependent restriction systems. Many key relevant discoveries were made by NEB scientists, who then genetically tailored E. coli strains to be tolerant of those restriction enzymes (2).
Cloning Vectors and Promoters: Those first efforts in cloning used the E. coli plasmid pBR322, an early vector developed by Francisco Bolivar and Ray Rodriguez, who were postdoctoral researchers in Herb Boyer’s laboratory at the University of California in San Francisco. Incidentally, it was Herb Boyer who discovered the EcoRI restriction enzyme and demonstrated that the “sticky” ends it created could join DNA fragments from different sources, making it the first restriction enzyme useful for DNA cloning.
NEB used derivatives of pBR322 that carried λPL (a powerful leftward promoter from Lambda bacteriophage), which is controlled by temperature: “off” at 32 °C and “on” at 42 °C. Because pBR322 has only a moderate copy number (~30–40 copies) per cell, the company quickly switched to a higher copy-number plasmid, pUC19, developed by Jo Messing at the University of California at Davis. That vector provided multiple cloning sites and a much higher copy number (~250 copies per cell), and it used a promoter from the lac operon.
In 1984, William Studier of Brookhaven National Laboratories developed an inducible T7 promoter system. With his method, a target gene is cloned downstream of the T7 promoter that is recognized by T7 RNA polymerase (from a gene integrated into the E. coli genome of engineered expression strains). This strong promoter system often can induce production of heterologous proteins comprising up to 50% of total cellular protein. This approach became popular both at NEB and throughout the field of biotechnology.
Our company’s internal efforts on recombinant restriction enzymes soon paid off. In 1982, PstI became the first product cloned and expressed by NEB scientists. A recombinant E. coli strain overexpressed about 100× more than the native organism. That allowed the company to reduce the unit price of PstI by 20-fold. Next, the company cloned, overexpressed, and sold an increasing number of restriction enzymes each year beginning with EcoRI, HaeII, and HindIII, then followed by many more. Today, nearly all of the >250 restriction enzymes it sells are purified from overexpression clones made by NEB.
Purification Using Affinity Chromatography
Soon after the company began producing recombinant restriction enzymes, the desire arose to couple efficient purification to the expression process. In the mid-1980s NEB began to research one of the first affinity-tagging systems, fusing the gene encoding E. coli’s maltose binding protein (MBP) in-frame with a target gene of interest. The resulting “fusion” protein then can be purified using amylose chromatography resin before the fusion tag is removed using a site-specific protease. This pMAL protein fusion and purification system was released in 1988 in the company’s first kit for protein expression and purification. Later it was discovered that MBP also has a natural ability to increase the solubility of fused target proteins significantly in E. coli.
In the following years, biotechnologists’ interest in affinity tags exploded. Additional fusion proteins were developed and used: e.g., glutathione S-transferase (GST), chitin-binding domain (CBD) and small peptide tags such as polyhistidines (poly-His), FLAG octapeptides, S-tags, streptavidin II, and polyarginines (poly-Arg). Of those, the most influential was poly-His tagging, which was developed by Roche in the late 1980s. His-tagged fusion proteins can be recovered using immobilized-metal affinity chromatography (IMAC), typically using Ni2+ beads or resins. To the present day, the combination of poly-His–tagged protein expression with IMAC is the most common approach to affinity-based nonantibody protein purification. The method tolerates a wide range of conditions, including the presence of protein denaturants, high salt concentrations, and detergents. It also can be used with many common cell-lysis reagents and a number of buffer additives.
Removal of affinity tags and/or fusion partners from purified recombinant proteins usually is achieved through digestion with a site-specific protease. A drawback to this approach is that a released target protein needs to be purified from the liberated tag and protease through additional chromatography steps. If a fusion partner happens to contain the same affinity tag as the protease, that can simplify purification of the target protein. An increasingly popular approach is to remove both the fusion partner (e.g., 6His–MBP) and the protease (e.g., His-tagged TEV protease) in a single IMAC capture step. This technique is used in the NEBExpress MBP fusion and purification system (Figure 2).
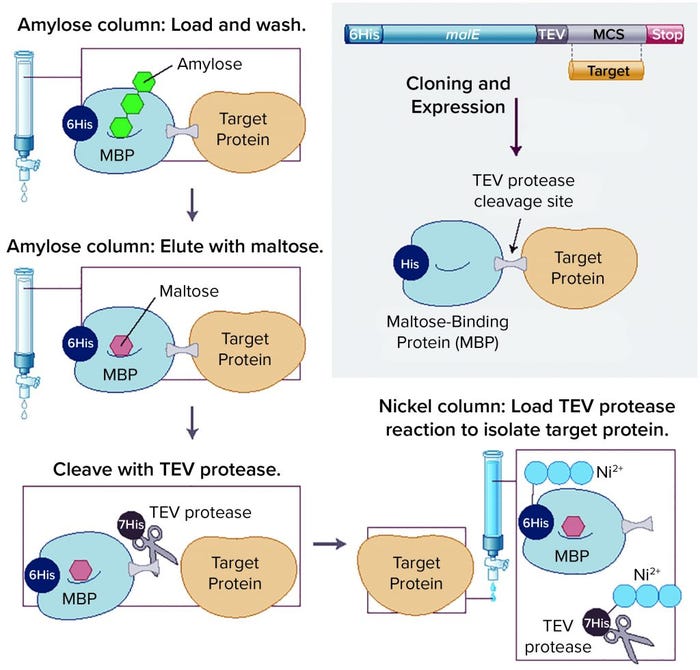
Figure 2: Overview of the NEBExpress MBP fusion and purification system (previously known as the pMAL protein fusion and purification system), in which a target protein is fused to maltose-binding protein (MBP) to enhance solubility and expression, followed by a simple purification technique; TEV = tobacco etch virus
Another NEB approach to affinity protein purification makes use of autosplicing protein domains called inteins. First described in 1990, they were shown to be a protein domain that can catalyze its own excision from a protein (3, 4). NEB researchers were studying inteins because of their presence in certain hyperthermophilic DNA polymerases and as a result were involved in elucidating the intein reaction mechanism (5). That research soon converged with other work in protein expression to yield a new intein-mediated strategy for fusion-protein removal without the need for protease-based cleavage (6).
In the new approach, E. coli expression of a target protein carrying an intein–chitin binding domain (intein-CBD) tag enables one-step purification using a chitin resin. When cell lysate passes over the resin, fusion proteins become immobilized; the target protein is released from CBD by inducing intein autocleavage through addition of a thiol-containing buffer or simply by a pH shift. NEB commercialized this work as the IMPACT (intein-mediated purification with an affinity chitin-binding tag) kit in the late 1990s (6).
Solving Protein Expression Problems
As the company has grown, so has its need to express classes of proteins aside from restriction enzymes. This development has presented new challenges because not all proteins express well (or at all) in E. coli. In addition to offering the familiar BL21 and BL21(DE3) expression strains, NEB has focused on solving expression of “difficult” proteins. Our scientists have sought to improve the ability of E. coli to express challenging proteins, including those with multiple disulfide bonds
and/or transmembrane domains and those that can be toxic to the host cells.
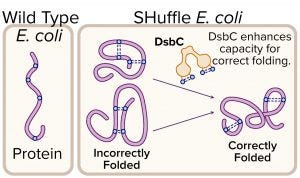
Figure 3: Expression of protein with multiple disulfide bonds using SHuffle competent Escherichia coli; disulfide bond formation in the cytoplasm of wild-type E. coli is not favorable, whereas SHuffle E. coli can fold such proteins correctly in their cytoplasm.
Proteins Containing Disulfide Bonds: Disulfide bonds are posttranslational covalent linkages formed by oxidation of a cysteine pair. Native disulfide bonds increase the stability of a protein, so they often are found in proteins that reside outside the chaperone-rich environment of the cytoplasm — e.g., secreted peptides, hormones, antibodies, interferons, and extracellular enzymes. When such proteins are expressed in E. coli cytoplasm, it can be difficult for them to fold correctly. In 2009, NEB commercialized SHuffle expression strains, which are engineered to support correct folding of proteins with multiple disulfide bonds (Figure 3). These strains constitutively express DsbC disulfide isomerase within their cytoplasm to promote correction of misoxidized proteins (7).
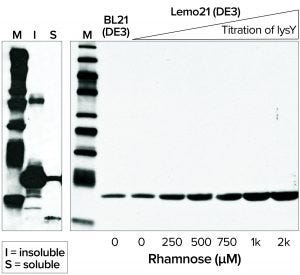
Figure 4: Western blot analysis of 6His-tagged Brugia malayi protein; (left) B. malayi protein expressed at 20 °C in BL21(DE3) competent E. coli; (right) soluble fractions of B. malayi protein expressed at 30 °C in BL21(DE3) or Lemo21(DE3) competent E. coli
Membrane or Toxic Protein Expression: Expression of membrane proteins is challenging for most heterologous systems, often resulting in protein aggregation and misfolding because transmembrane segments are hydrophobic. When working with E. coli as a host, it is advantageous to express such proteins in moderation and thus prevent saturation of the membrane protein biogenesis pathway. NEB’s Lemo21(DE3) competent E. coli strain was designed for tunable expression to achieve optimal assembly of transmembrane proteins and optimal folding of soluble proteins (Figure 4) (8).
For cases in which heterologous proteins are toxic to host cells, tightly controlling gene expression can improve host viability by maintaining expression levels of a toxic target protein just below the host strain’s tolerance. In strong T7-promoter–based systems, an effective means of controlling expression is to use a host strain that expresses a T7 RNA polymerase inhibitor protein (LysY), such as in NEB’s Lemo21(DE3) or T7 Express lysY/Iq strains.
To express a highly toxic protein, it may be necessary to use a cell-free expression system. NEB’s PURExpress in vitro protein synthesis kit is reconstituted from purified components necessary for E. coli translation. This kit also can be used with PURExpress disulfide bond enhancers to improve protein folding. An alternative is the NEBExpress cell-free E. coli protein synthesis system using a cell lysate to provide high-level expression of target proteins from linear or plasmid DNA templates. (For more information on cell-free expression, see the article by B. Melinek et al. elsewhere in this issue.)
The Future of Protein Expression
The biotechnology field of protein expression is constantly evolving. Applications such as protein engineering and synthetic biology are driving advancement toward high-throughput protein expression. Researchers now want to test hundreds, even thousands, of expressed proteins in a single day — to narrow their focus quickly to the most interesting variants. For traditional cloning methods, introducing vectors into a host strain for cell propagation takes multiple days. So cell-free protein expression (which can be accomplished in an hour or less) will become increasingly important in the coming years.
Just as in vivo protein expression came from humble beginnings and has progressed to bring about highly engineered host strains and regimented bioprocessing, we anticipate a similar revolution in cell-free protein expression systems. A new generation of NEB scientists is dedicated to advancing cell-free expression by engineering novel cell lines as extract sources, developing improved cell-free manufacturing processes (e.g., PURExpress or NEBExpress technologies), optimizing cell-free system formulations, and exploring the potential for system scale up to produce milligram to gram quantities of proteins without cell culture.
References
1 Rosano GL, Ceccarelli EA. Recombinant Protein Expression in Microbial Systems. Front. Microbiol. 5, July 2014: 172; https://doi.org/10.3389/fmicb.2014.00341.
2 Raleigh EA, Wilson G. Escherichia coli K-12 Restricts DNA Containing 5-Methylcytosine. PNAS 83(23) 1986: 9070–9074; https://doi.org/10.1073/pnas.83.23.9070.
3 Hirata R, et al. Molecular Structure of a Gene, VMA1, Encoding the Catalytic Subunit of H(+)-Translocating Adenosine Triphosphatase from Vacuolar Membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265(12) 1990: 6726–6733.
4 Kane PM, et al. Protein Splicing Converts the Yeast TFP1 Gene Product to the 69-kD Subunit of the Vacuolar H(+)-Adenosine Triphosphatase. Science 250(4981) 1990: 651–657; https://doi.org/ 10.1126/science.2146742.
5 Perler FB, Xu MQ, Paulus H. Protein Splicing and Autoproteolysis Mechanisms. Curr. Opin. Chem. Biol. 1(3) 1997: 292–299; https://doi.org/10.1016/s1367-5931(97)80065-8.
6 Chong S, et al. Single-Column Purification of Free Recombinant Proteins Using a Self-Cleavable Affinity Tag Derived from a Protein Splicing Element. Gene 192(2) 1997: 271–281. https://doi.org/10.1016/s0378-1119(97)00105-4.
7 Lobstein J, et al. SHuffle, a Novel Escherichia coli Protein Expression Strain Capable of Correctly Folding Disulfide Bonded Proteins in Its Cytoplasm. Microb. Cell. Fact. 11, May 2012: 56; https://doi.org/10.1186/1475-2859-11-56.
8 Wagner S, et al. Tuning Escherichia coli for Membrane Protein Overexpression. PNAS 105(38) 2008: 14371–14376; https://doi.org/10.1073/pnas.0804090105.
Christopher H. Taron, PhD, is scientific director and James C. Samuelson, PhD, is a senior scientist in the protein expression and modification division; and Lydia Morrison, MS, is a marketing communication writer and social media manager at New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938-2723; 1-978-927-5054; fax 1-978-921-1350; www.neb.com. New England Biolabs, NEB, pMAL, IMPACT, SHuffle, PURExpress, and NEBExpress are registered trademarks.
You May Also Like






