Capture of CH1-Containing Bispecific Antibodies: Evaluating an Alternative to Protein ACapture of CH1-Containing Bispecific Antibodies: Evaluating an Alternative to Protein A
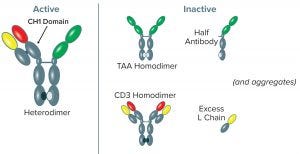
Figure 1: Variant forms of BsAb CD3-TAA–expressed products; the CD3-TAA heterodimer (containing one light (L) and two heavy (H) chains) is the active form. Other forms include the TAA homodimer, CD3 homodimer, half-antibody, and L chain, all of which are inactive. Aggregated forms of product also are present. Note that only the full-length anti-CD3 human heavy chain has a CH1 domain, and the anti-CD3 human heavy chain is present in only two forms: the CD3-TAA heterodimer and CD3 homodimer.
Bispecific antibodies (BsAbs) are designed to recognize and bind two different antigens, in many cases for the purpose of immune effector-cell activation to destroy cancer cells (1). Such BsAbs mediate cell killing by binding simultaneously to an antigen that is overexpressed on tumor cells and to the CD3 receptor, activating cytotoxic T lymphocytes (2). Using proprietary UniRat human heavy-chain technology combined with OmniFlic human fixed–light-chain antibody technology licensed from Ligand Pharmaceuticals, Teneobio has produced several bispecific antibodies, each targeting a different tumor antigen and CD3 (3).
The lead molecules are composed of three chains: two nonidentical human heavy chains and a human kappa light chain. One heavy chain is full length (OmniFlic derived) and the other — discovered in UniRat transgenic rats (4) — is truncated, lacking the CH1 domain. The human kappa light chain is bound to the full-length heavy chain. Correct pairing of heavy chains is achieved through knobs-into-holes technology. It relies on noncovalent interactions, the result of complementary mutations in the CH3 domains (5) and hinge-region disulfide bonds, to form heterodimers (6).
The number of bispecific antibodies in clinical testing is growing rapidly (7), and so is the need for efficient production and purification strategies. The main challenge of producing bispecific antibodies comes with their heterogenous nature. It brings an inherent potential to form inactive product variants that copurify with active products. That makes purification critical. Figure 1 illustrates the variants of bispecific antibody CD3-TAA–expressed products. The predominant active form is a heterodimer (BsAb CD3-TAA); however, numerous inactive forms consisting of CD3 and TAA homodimers, half-antibodies, and excess light chains can contaminate expressed product.
The common method for purifying monoclonal antibodies (MAbs) and BsAbs following harvest and clarification uses affinity capture with a commercially available protein A resin. Although used for bulk capture and high-resolution binding of antibody products, the method can be challenging with bispecific antibodies because of the multiplicity of structures. Teneobio faced two potential issues when considering protein A as the method of capture: First, all variants containing intact Fc domains (BsAb CD3-TAA, CD3 homodimer, and TAA homodimer) would copurify. Second, the low-pH condition required for elution from protein A potentially causes aggregation of both active and inactive BsAb product variants (8).
Methods to reduce the risk of aggregation during protein A chromatography have been discussed elsewhere. Most studies have focused on moderating the pH needed for elution. For example, 2 M arginine at pH 4.3 has been used to elute human human immunoglobulin G (IgG) 1 from protein A under relatively mild conditions (9). An alternative approach makes use of novel ligands for affinity capture. A purification matrix based on the ZCa protein A–derived domain displays calcium-dependent binding of IgG and elution under mild conditions (10).
We investigated using commercially available CaptureSelect CH1-XL affinity resin from Thermo Fisher Scientific for purification of bispecific antibody CD3-TAA. The resin is based on a 13-kDa llama VHH antibody fragment immobilized on epoxide-activated agarose (65 µm) that binds specifically to the CH1 domain of IgG heavy chain. Originally developed for purifying human Fab fragments (11–13), the ligand does not bind to often-overexpressed light chains and light-chain dimers, it recognizes all four human IgG subclasses, and it elutes under mild conditions. Although this resin was not marketed initially for purification of bispecific antibodies, we noted that its selectivity for human antibodies and derivatives containing the CH1 domain makes it potentially well suited to the purification challenges of BsAb CD3-TAA. CaptureSelect CH1-XL resin binds preferentially to the heterodimeric BsAb CD3-TAA product and its CD3 homodimer, but not to the TAA homodimer, making it a particularly good choice for our application.
Presence of the CD3 homodimer constitutes a safety risk from potential nonspecific activation of T cells in vivo (14). Further, because CD3 homodimer contains CH1 domain, similarity with the BsAb makes it difficult to remove the homodimer during purification. Therefore, we sought to minimize production of the knob–knob CD3 homodimer during expression. To achieve that, we constructed each individual gene on a separate plasmid. Then we tested different plasmid ratios to optimize relative polypeptide chain expression and found that excess TAA‑Fc plasmid DNA is required over CD3-encoding plasmid. With that approach, we optimized harvest compositions to contain <5% of the undesired CD3 homodimer while maintaining high titers of bispecific antibodies.
The TAA homodimer is the second most prominent molecular species. The unique selectivity of CaptureSelect CH1‑XL resin also enables efficient removal of this homodimer, which lacks a CH1 domain, while capturing the desired heterodimer BsAb CD3-TAA.
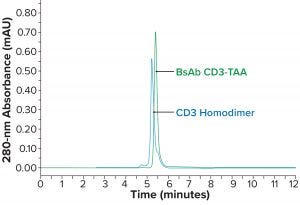
Figure 2a: Comparing BsAb CD3-TAA and homodimers; SEC-HPLC of active heterodimer BsAb CD3-TAA and CD3 homodimer (similar variants, with the CD3 homodimer appearing to be slightly larger in size)
Materials and Methods
Materials: All ultrapure and reagent-grade chemicals came from commercial sources. In-house BsAb CD3-TAA was expressed by Chinese hamster ovary (CHO) cells. Harvested cell culture fluid (HCCF) was the feedstock for these purification experiments.
BsAb CD3-TAA Affinity Purification: Purification of BsAb CD3-TAA using MabSelect SuRe XL resin (Cytiva, formerly GE Healthcare) was performed according to the manufacturer’s protocol. Briefly, 50 mL of HCCF were loaded onto a 1-mL MabSelect Sure XL HiTrap column (7 × 25 mm) equilibrated in 50 mM Tris at pH 7.0. After loading, the column was washed with equilibration buffer, then bound product was eluted with 25 mM citric acid at pH 3.6. Eluted pools were neutralized using 1 M Tris at pH 9.0.
For CaptureSelect CH1-XL affinity chromatography, 50 mL of HCCF were loaded onto a 9-mL CaptureSelect CH1‑XL column (16 × 45 mm) equilibrated in 50 mM Tris buffer at pH 7.0. After loading, the column was washed with 50 mM Tris and 0.5 M NaCl at pH 7.0. Bound product was eluted with 50 mM acetic acid, 10% glycerol, and 10% sucrose at pH 4.0. Eluted pools were neutralized using 1 M Tris at pH 9.0.
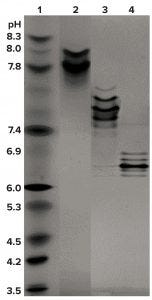
Figure 2b: Comparing BsAb CD3-TAA and homodimers; IEF of CD3 homodimer (lane 2, pI 7.6–8.0), BsAb CD3-TAA heterodimer (lane 3, pI 7.4–7.6), and TAA homodimer (lane 4, pI 6.2–6.9); pI markers are in lane 1.
Size-Exclusion Chromatography: To determine purity of the BsAb mixture, an UltiMate 3000 ultrahigh-pressure liquid chromatograph (UHPLC) from Thermo Fisher with a TSKgel UP-SW3000 column (Tosoh Bioscience) was used to perform size-exclusion chromatography. The mobile phase was 100 mM sodium citrate, 500 mM NaCl, and 200 mM l-arginine at pH 6.2. The column was equilibrated with that mobile phase and checked for a stable baseline. Flow rate was set at 0.25 mL/min in isocratic mode, with an injection amount of 50 µg and a run time of 10 minutes. Detector wavelengths were set at 280 nm and 220 nm. Purified BsAb CD3-TAA was used as a reference standard.
Isoelectric Focusing (IEF): To characterize variants by charge, we ran a pH 3-10 IEF gel (Thermo Fisher Scientific), having used IEF markers 3–10 from SERVA to calibrate the gel. A pH gradient was developed using the following protocol:
1 hour, 100 V, 18 mA, 2 W
1 hour, 200 V, 18 mA, 3.5 W
0.5 hour, 500 V, 18 mA, 9 W.
IEF bands were visualized using InstantBlue stain from Expedeon.
Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE): We investigated the purity of our BsAb samples using an X-Cell SureLock Mini-Cell system with NuPage 4–12% Bis Tris gels in Invitrogen MES SDS running buffer (Thermo Fisher Scientific). Samples were treated with βME and 4× SDS sample buffer (also Invitrogen) then heated before being loaded (2 µg/lane). Gels were run at a constant 200 V for 35 minutes, then stained for 10–30 minutes using InstantBlue gel stain (Expedeon) and destained for a total of 20 minutes with deionized water. We imaged the gels using the GelDoc system (Bio-Rad Laboratories) and compared sample sizes using PageRuler prestained standard (Thermo Fisher Scientific).
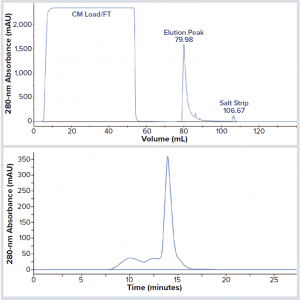
Figure 3: BsAb CD3-TAA aggregates following elution from MabSelect Sure XL protein A resin; (top) chromatogram of BsAb CD3-TAA purified from harvested cell culture fluid (HCCF) on protein A and eluted at pH 3.6; (bottom) analysis by SEC-HPLC of the protein A elution peak. The major peak (~14 minutes) represents monomer IgG (72%); earlier eluting peaks (~10–12 minutes) represent high–molecular-weight (HMW) aggregated forms (28%).
Results
Isoelectric focusing and size-exclusion high-performance liquid chromatography (SEC-HPLC) were used for initial characterization of the BsAb mixture. We found that the active heterodimer of BsAb CD3-TAA is similar in size to both the inactive CD3 (Figure 2a) and TAA (data not shown) homodimers. Although the sizes of these three oligomers are similar, they do have distinctly different isoelectric points (pIs) (Figure 2b).
In our first attempts to capture and purify BsAb CD3-TAA, we used commercially available protein A resin. However, the pH sensitivity of BsAb CD3-TAA created challenges using the acidic conditions required for efficient elution from protein A (pH <3.6). Figure 3, top shows an elution profile at pH 3.6 of BsAb CD3-TAA after protein A capture. That elution was efficient. However, the elution peak collected and analyzed (Figure 3, bottom) by SEC-HPLC shows that eluted product is significantly aggregated.
As expected, because all three variants contain intact Fc domains, the homodimer variants copurify with BsAb CD3-TAA (data not shown). We attempted to elute the product from protein A using milder conditions, but those attempts had limited success due to incomplete product recovery (data not shown).

Figure 4: BsAb CD3-TAA shows much reduced aggregation on elution from CaptureSelect CH1 XL resin; (left) chromatogram of BsAb CD3-TAA purified from harvested cell culture fluid on the resin and eluted at pH 4.0; (right) analysis by SEC-HPLC of the elution peak. Monomer IgG (11.115 min) is 98%; high–molecular-weight (HMW) forms (9.673 min) are 2%.
To address the shortcomings of protein A for this application, we identified and tested an affinity medium with a different ligand and unique purification properties. CaptureSelect CH1-XL resin binds to the CH1 domain of human IgG with a nanobody ligand that is unrelated to Staphyloccocus aureus protein A. The resin is designed to bind tightly to human IgG (with low nanomolar affinity) at neutral pH. However, that affinity is reduced at mildly acidic pH, enabling elution under less acidic conditions than protein A requires. Given the aggregation issues that we faced when using the harsh elution condition required for protein A, we tested elution of BsAb CD3-TAA from CaptureSelect CH1-XL resin in 50 mM acetic acid, 10% glycerol, and 10% sucrose at pH 4 and found that it provided for efficient elution in two column volumes (Figure 4, left). To determine the extent of aggregation, we analyzed the BsAb CD3-TAA eluate with SEC-HPLC (Figure 4, right). Monomeric BsAb CD3-TAA and high–molecular-weight (HMW) peaks were determined to be 98% and 2%, respectively, showing reduced aggregation compared with material eluted from protein A.
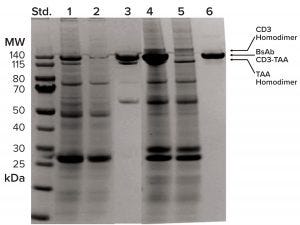
Figure 5: TAA homodimer is separated from BsAb CD3-TAA by CaptureSelect CH1 XL resin but not by protein A. SDS-PAGE analysis of chromatography fractions shows column performance in lanes 1–3 for protein A and lanes 4–6 for CaptureSelect CH1 XL; left (standard) lane shows molecular-weight standards; lanes 1–3 show protein A load, flow-through, and elution fractions, respectively; lanes 4–6 show CaptureSelect CH1 XL load, flow-through, and elution fractions, respectively. Load and flow-through fractions are 10 µL each; pool fractions are 2 µg each. Arrows indicate positions of BsAb CD3-TAA and homodimers.
In addition to the potential for milder elution conditions, CaptureSelect CH1-XL resin also provides excellent selectivity for our BsAb application. By contrast with protein A, which captures all Fc-containing molecular variants, this resin captures only CH1-domain–containing variants (BsAb CD3-TAA and CD3 homodimer). The TAA homodimer lacking a CH1 domain is expected to flow through the column. During purification development, we confirmed that higher product purity could be achieved using this resin. Figure 5 compares a stained SDS-PAGE gel that compares the results of protein A (lanes 1–3) and CaptureSelect CH1-XL (lanes 4–6) capture. The elution pool derived from protein A (lane 3) contains both BsAb CD3-TAA and TAA homodimer. By contrast, we achieved high purity of BsAb CD3-TAA in the CaptureSelect CH1-XL pool (lane 6), with both homodimers clearly present in the column flow-through fraction (lane 5).
We also measured the dynamic binding capacity (DBC) of BsAb CD3-TAA on CaptureSelect CH1-XL resin by loading 1-mL CaptureSelect CH1-XL columns (0.7 × 2.5 cm) with purified BsAb CD3-TAA (5 mg/mL) at each of four flow rates to achieve residence times of one, two, four, and eight minutes. Column eluate was detected using optical density (OD) of 280 nm to measure 10% breakthrough. In agreement with the manufacturer’s recommendation, we determined that the DBC increased directly with residence time. Maximum DBC (20 mg/mL of BsAb) is reached at four minutes (data not shown).
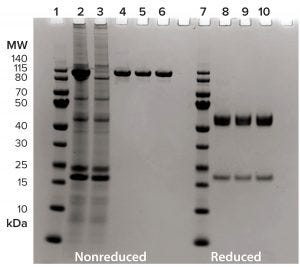
Figure 6: Summary of BsAb CD3-TAA purification through downstream process using 4–12% SDS-PAGE, 2 µg/lane, and InstantBlue stain. Lanes 1–6 are nonreduced; lanes 7–10 are under reducing conditions. Lanes 1 and 7 are molecular-weight standards; lane 2 is harvested cell culture fluid; lane 3 is the CH1 XL flow-through fraction; lanes 4 and 8 are from the CH1 XL elution pool; lanes 5 and 9 are from the intermediate purification-step pool; and lanes 6 and 10 are from the polishing purification-step pool.
Based on the superior performance of CaptureSelect CH1-XL resin for capture of BsAb CD3-TAA, we proceeded to design a downstream process that includes additional chromatography steps for purification. Figure 6 summarizes the complete process for BsAb CD3-TAA. The resin’s selectivity for our product is apparent (lanes 2–4), as is the ability of the purification scheme to yield a clean product (lane 6).
Discussion
Whereas the utility of protein A affinity chromatography for capture of MAbs is appreciated widely and forms the basis for antibody downstream processes worldwide, its usefulness for capture of antibody derivatives such as Fc-containing bispecific molecules is not as straightforward. Indeed, protein A use is complicated by the fact that antibody derivatives often are expressed in mixtures containing some product variants that have identical binding affinity to protein A. Moreover, the need for low-pH (<3.6) elution in protein A chromatography often can be incompatible with engineered protein structures that are prone to aggregate under harsh conditions. For those reasons, we decided to test CaptureSelect CH1-XL resin for affinity capture of our bispecific construct (which contains a full-length human heavy chain) and found that it successfully addressed both issues.
Elution was achieved under relatively mild conditions (pH 4.0). To further reduce the potential of BsAb CD3-TAA to aggregate, we included glycerol and sucrose (10% each) as stabilizers in the elution buffer. Such polyols are known to protect proteins from unfolding under stressful conditions (15). The acid lability of BsAb CD3-TAA not only created issues for protein A capture, but it also prevented the use of a low-pH hold for virus inactivation as a subsequent step in the process. Consequently, to adapt the downstream process further, we implemented a Triton X100 detergent-based method for virus inactivation (16).
We found CaptureSelect CH1-XL resin DBC to be lower than that of protein A. The manufacturer reports a DBC of 19 mg for polyclonal human Fab/mL at two minutes residence time. In our study, however, we determined the DBC for our bispecific antibody CD3-TAA to be 20 mg/mL at four minutes residence time. Because Fab represents ~40% of the mass of BsAb CD3-TAA, the predicted DBC should be higher than what we observed (e.g., ~46 mg/mL of a 120-kDa BsAb). It is possible that the larger BsAb sterically hinders some binding sites and thus reduces overall binding capacity. Indeed, the manufacturer reports that ligand use is lower for IgGs than for Fab fragments, resulting in a lower molar DBC.
Although the value of an affinity step for purifying product out of crude feedstock is clear, removal of homodimers and other variants in bispecific mixtures also can be achieved using nonaffinity chromatographic methods. Tang et al. recently reported successful removal of half-antibodies, homodimers, and aggregates during bispecific antibody purification with Cytiva’s Capto MMC ImpRes mixed-mode chromatography (17). So that method might be considered as a polishing step in a purification scheme for removing homodimers and other variants that remain in bispecific mixtures.
The presence of product-related impurities in BsAb preparations presents a unique purification challenge compared with downstream process development for MAbs. Capture chromatography using protein A, which has enabled establishment of platform downstream processes for MAbs in a way that no other purification tool could have done, cannot be applied readily to bispecific mixtures. The growing importance of engineered bispecific antibodies as a therapeutic class offers a new opportunity for downstream processes to be designed around alternative affinity-capture ligands.
Acknowledgments
We are grateful to Chris Rooney for assistance with graphics and Wendy Lin and Frank Detmers for critical review of our manuscript.
References
1 Labrijn AF, et al. Bispecific Antibodies: A Mechanistic Review of the Pipeline. Nature Rev. Drug Disc. 18(8) 2019: 585–608; https://doi.org/10.1038/s41573-019-0028-1.
2 Sedykh SE, et al. Bispecific Antibodies: Design, Therapy, Perspectives. Drug Des. Devel. Ther. 12, 2018: 195–208; https://doi.org/10.2147/DDDT.S151282.
3 Harris KE, et al. Sequence-Based Discovery Demonstrates That Fixed Light Chain Human Transgenic Rats Produce a Diverse Repertoire of Antigen-Specific Antibodies. Front. Immunol. 9, 2018: 889; https://doi.org/10.3389/fimmu.2018.00889.
4 Clarke SC, et al. Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies. Front. Immunol. 9, 2018: 3037; https://doi.org/10.3389/fimmu.2018.03037.
5 Ridgway JB, Presta LG, Carter P. “Knobs-Into-Holes” Engineering of Antibody CH3 Domains for Heavy Chain Heterodimerization. Protein Eng. 9(7) 1996: 617–621; https://doi.org/10.1093/protein/9.7.617.
6 Shatz W, et al. Knobs-into-Holes Antibody Production in Mammalian Cell Lines Reveals That Asymmetric Afucosylation Is Sufficient for Full Antibody-Dependent Cellular Cytotoxicity. MAbs 5(6) 2013: 872–881; https://doi.org/10.4161/mabs.26307.
7 Suurs FV, et al. A Review of Bispecific Antibodies and Antibody Constructs in Oncology and Clinical Challenges. Pharmacol. Ther. 201, 2019: 103–119; https://doi.org/10.1016/j.pharmthera.2019.04.006.
8 Tustian AD, et al. Development of a Novel Affinity Chromatography Resin for Platform Purification of Bispecific Antibodies with Modified Protein A Binding Avidity. Biotechnol. Prog. 34(3) 2018: 650–658; https://doi.org/10.1002/btpr.2622.
9 Arakawa T, et al. Elution of Antibodies from a Protein-A Column By Aqueous Arginine Solutions. Protein Expr. Purif. 36(2) 2004: 244–248; https://doi.org/10.1016/j.pep.2004.04.009.
10 Scheffel J, et al. Optimization of a Calcium-Dependent Protein A–Derived Domain for Mild Antibody Purification. MAbs 11(8) 2019: 1492–1501; https://doi.org/10.1080/19420862.2019.1662690.
11 Eifler N, et al. Development of a Novel Affinity Chromatography Resin for Platform Purification of Lambda Fabs. Biotechnol. Prog. 30(6) 2014: 1311–1318; https://doi.org/10.1002/btpr.1958.
12 Spooner J, et al. Evaluation of Strategies to Control Fab Light Chain Dimer During Mammalian Expression and Purification: A Universal One-Step Process for Purification of Correctly Assembled Fab. Biotechnol. Bioeng. 112(7) 2015: 1472–1477; https://doi.org/10.1002/bit.25550.
13 Detmers FG, et al. Rapid Implementation of Novel Affinity Purification: Manufacture of Commercial-Scale Next-Generation Antibody Therapies. BioProcess Int. 17(10) 2019: 12–17; https://bioprocessintl.com/sponsored-content/rapid-implementation-of-novel-affinity-purification-manufacture-of-commercial-scale-next-generation-antibody-therapies.
14 Minguet S, et al. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity 26(1) 2007: 43–54; https://doi.org/10.1016/j.immuni.2006.10.019.
15 Back JF, Oakenfull D, Smith MB. Increased Thermal Stability of Proteins in the Presence of Sugars and Polyols. Biochemistry 18(23) 1979: 5191–5196; https://doi.org/10.1021/bi00590a025.
16 Roberts PL. Virus Inactivation By Solvent/Detergent Treatment Using Triton X-100 in a High Purity Factor VIII. Biologicals 36(5) 2008: 330–335; https://doi.org/10.1016/j.biologicals.2008.06.002.
17 Tang J, et al. Removal of Half Antibody, Hole–Hole Homodimer, and Aggregates During Bispecific Antibody Purification Using MMC ImpRes Mixed-Mode Chromatography. Protein Expr. Purif. 167, 2020: 105529; https://doi.org/10.1016/j.pep.2019.105529.
Corresponding author Steven M. Chamow is founder and principal consultant, and Angela L. Linderholm is an associate at Chamow and Associates, Inc., San Mateo, CA; [email protected]. Katherine E. Harris is senior director of discovery, Ute Schellenberger is vice president of protein science, and Brett Jorgensen is director of process development at Teneobio, Inc., Newark, CA. Payal P. Pratap is a graduate student at Scripps Research Institute, La Jolla, CA.
OmniFlic is a registered trademark of Ligand Pharmaceuticals, Inc. UniRat is a trademark of Teneobio, Inc. CaptureSelect, PageRuler, UltiMate, and XCell SureLock are trademarks of Thermo Scientific Inc. TSKgel is a registered trademark of Tosoh Bioscience. InstantBlue is a registered trademark of Expedion Ltd. Gel Doc is a trademark of Bio-Rad Laboratories. MabSelect SuRe and Capto MMC ImpRes are trademarks of Cytiva (formerly GE Healthcare). Triton is a trademark of Dow Chemical Company.
You May Also Like