Evaluating Adsorptive Filtration As a Unit Operation for Virus Removal February 2015Evaluating Adsorptive Filtration As a Unit Operation for Virus Removal February 2015
To date, the majority of recombinant monoclonal antibodies (MAbs) have been produced by mammalian cells. During such production processes, the potential risk of entrained viruses must be critically considered (1). Contamination can arise from animal cell lines or from adventitious viruses introduced during manufacturing. To ensure the viral safety of biotechnology products, companies can take four complementary approaches (2, 3):
Using animal-component–free raw materials wherever possible
Virus testing of master cell banks
Virus testing of unprocessed harvest
Performing downscale virus validation studies on selected process steps to demonstrate the capacity of the process to remove and inactivate viruses.

In most antibody-purification processes, the first viral clearance step is a low-pH inactivation (pH <4) conducted after protein A affinity capture chromatography. Precipitation of impurities often occurs in this low- pH treatment and subsequent conditioning, and filtration is required afterward before further processing in chromatographic polishing steps. Depth filters are commonly used (Figure 1).
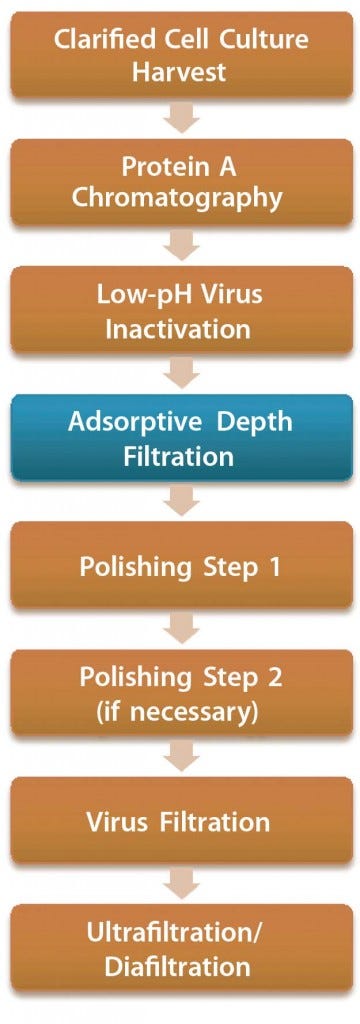
Figure 1: Antibody purification process with adsorptive depth filtration as a viral clearance step orthogonal to low-pH inactivation and virus filtration; the additional virus removal step in this process presumably enables replacement of anion-exchange chromatography as a polishing step while maintaining the virus safety of the process. Thus, a two-column purification process could be implemented (4, 9).
Many processors apply charged depth filters to make better use of that filtration step. Charged depth filtration is a scalable, well- characterized unit operation commonly used in blood-plasma fractionation processes, providing viral log clearance of ≤6 LRV (log10 reduction value) (7, 8).
Previous investigations of antibody processes have demonstrated the ability of positively charged depth filters to reduce host-cell proteins (HCPs) and DNA (4) and to effectively remove viruses (1, 5, 6). Some investigators postulate that the virus retention can be attributed to anionic adsorptive effects of mammalian viruses on positively charged filter media (5). Such recommended model viruses for downscale virus validation studies as xenotropic murine leukemia virus (X-MuLV) and minute virus of mice (MVM) have a negative net charge at neutral or basic pH. Consequently, they bind to the positively charged surface of depth filters.
Our objective was to demonstrate the capabilities of three different adsorptive filters to remove viral contamination under typical process conditions. We tested laboratory-scale modules of these 3M filter types: Zeta Plus EXT 60ZA05A, Zeta Plus EXT 90ZA05A, and the functionalized nonwoven (FNW) Emphaze anion- exchange (AEX) Hybrid Purifier. The chromatographic characteristic of the latter filter comes mainly from its comparably high content of positively charged quaternary groups captured within a non-woven matrix. For these downscale virus retention studies, we used the mammalian viruses X-MuLV and MVM.
Materials and Methods
For these adsorptive depth filtration studies, we used ÄKTA Explorer 10 systems with Unicorn software from GE Healthcare. All buffers were 0.2-µm filtered before use. Table 1 lists our adsorptive test filters with anionic exchange functionality.
Load Material and Virus Assay: Rentschler scientists performed virus- spiking experiments at Charles River Biologics Testing Solutions in Cologne, Germany, using X-MuLV and MVM as model viruses (Table 2). Challenge materials were prepared from processed MAb-containing harvest.
Table 1: Overview of used filter types and process parameters used
Zeta Plus EXT 60ZA05A | Zeta Plus EXT 90ZA05A | Emphaze AEX FNW | |
|---|---|---|---|
Material | Cellulose-fiber | Cellulose-fiber | Charge-modified, functionalized nonwoven |
Filter type | Disc | Disc | Disc |
Charge | Positive | Positive | Positive |
Filter layers | 2 | 2 | 4 |
Cross-sectional area | 3.8 cm² | 3.8 cm² | 3.8 cm² |
Nominal pore size | 0.4–0.8 µm | 0.2–0.8 µm | 0.2–0.8 µm |
Used filter holder | Ligaster 25 EXT | Ligaster 25 EXT | Ligaster 25 EXT |
Table 2: Virus models used for the virus clearance study
X-MuLV | MVM | |
|---|---|---|
Family | Retrovirus | Parvovirus |
Size (nm) | 80 – 100 nm | 20 – 25 nm |
Lipid envelope | Yes | No |
Genome | ssRNA | ssDNA |
Physicochemical resistance | low | very high |
Rationale | Representative nondefective C-type retrovirus | C-type retrovirusModel for both human and animal parvoviruses |
Detector cell line | PG-4 | A9 |
Culture harvest was clarified using a 3M Zeta Plus encapsulated 16EZA system with E16E01A60SP02A capsules (3 × 0.23 m²), 0.2-µm filtered and purified using protein A affinity chromatography. Column elution was achieved using a phosphate–citrate– Tris buffer system, and the eluate was adjusted to pH 3.5 and diluted to a MAb concentration of 5 g/L. Before the adsorptive depth filtration runs, the protein solution was divided and each sample load adjusted to desired loading conditions (pH value and sodium chloride concentration) (Table 3). Those adjustments were made using a phosphate–citrate–1 M Tris buffer and 4M NaCl solution, leading to a final MAb concentration of 3.75 g/L.
Table 3: Experiments carried out with all three depth filters
pH 5.5 Low Salt | pH 7.0 Low Salt | pH 7.0 High Salt | |
|---|---|---|---|
X-MuLV | X | X | X |
MVM | X | X |
To exclude influence of test items on cell growth or virus replication, Charles River Biologics Testing Solutions also performed cytotoxicity and viral interference assays. The same test facility also provided virus stock solutions and further analyzed samples using 50% tissue-culture infective dose (TCID50) assays. The detection limit of such assays depends on the volume of sample that is incubated with indicator cells. Those cells were cultivated for a specific incubation period and inspected microscopically for virus-induced changes in their morphology.
Experimental Methods: Viral clearance experiments were performed with the three adsorptive filter types using three different buffer conditions, each in duplicate (Table 3). Virus- reduction factors of X-MuLV and MVM were determined for these conditions
phosphate–citrate–Tris buffer at pH 5.5 with 0 M NaCl (“pH 5.5 low salt”)
phosphate–citrate–Tris buffer at pH 7.0 with 0 M NaCl (“pH 7.0 low salt”).
Those buffers represent appropriate conditions for further processing in a MAb purification. Additionally, viral reduction of X-MuLV (80–100 nm) was determined in a phosphate–citrate–Tris buffer at pH 7.0 with 1M NaCl (“pH 7.0 high salt”) to minimize ionic interactions between viruses and the adsorptive filter material. By subtracting the high-salt condition’s virus LRV from that of the low-salt condition, we calculated the viral clearance related to adsorption. Because MVM (20–25 nm) is much smaller than the nominal pore size of these filter media (0.2–0.8 µm for the Zeta Plus VR series), no additional filtrations were run under high-salt conditions. We excluded the mechanical effects for retention of MVM.
Duplicate filtrations were run in parallel with two chromatography systems, so 180 mL of the affinity- purified and conditioned MAb solutions were spiked with 5% (v/v) (9 mL) of nonconcentrated, ultracentrifuged, and prefiltered virus stock solution (0.45-µm filter for X-MuLV and 0.1-µm filter for MVM). After mixing, a sample was withdrawn from the spiked pool and immediately analyzed for the virus titer (“load sample”). For X-MuLV, a second load sample was stored until the end of the filtration (“hold sample”). MVM is known to be physicochemically resistant (1).
Analysts loaded 220 L/m² filter area (84 mL) of the starting material at 158 L/m²/h (1 mL/min) to the equilibrated adsorptive depth filters. Subsequently, they flushed those filters with 63 L/m² (24 mL) equilibration buffer. At the end, they analyzed the total filtrate virus titer (both flow-through and flush, referred to herein as the “filtrate sample”) and determined the titer of the drawn “hold sample.” They calculated virus log-reduction values as described in Equation 1:
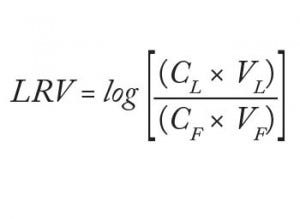
in which CL = virus concentration of the load, VL = volume of the load, CF = virus concentration in the filtrate, and VF = volume of the filtrate.
Results and Discussion
From cytotoxicity and viral- interference assays performed by Charles River Biologics Testing Solutions (data not shown), we obtained confirmation that no test items influenced cell growth or virus replication. The analysts also determined suitable sample dilutions for performing the TCID50 assays.
Removal of X-MuLV: Under low-salt conditions at pH 5.5 and pH 7.0, we observed no positive indicator cells of X-MuLV (TCID50 assay) in filtrate fractions from all three tested filters. Excellent virus removal was obtained, at least in the range of 5.0–5.8 LRV (Table 4). By contrast, under high-salt conditions at pH 7.0, we determined virus removal of 1.2 LRV (Emphaze AEX FNW) and 2.7 LRV (Zeta Plus 60ZA05A and 90ZA05A) filters. Because high-salt concentrations suppress electrostatic interactions, we used such conditions to investigate virus removal without attractive electrostatic interactions. The low value for the Emphaze AEX FNW filter shows no significant virus removal capacity at high-salt condition; the assay’s standard deviation is ±1 LRV.
Table 4: X-MuLV clearance results for the three charged depth filters tested
Filter | Zeta Plus 60ZA05A | Zeta Plus 90ZA05A | Emphaze AEX FNW |
|---|---|---|---|
Conditions | pH 5.5 and 0 M NaCl (low salt) | ||
Minimum reduction value (log10) | ≥5.84 | ≥5.66 | ≥5.84 |
Reduction value (log10) | ≥5.84 ± 0.23 | ≥5.66 ± 0.31 | ≥5.84 ± 0.28 |
Total virus load: load (log10) | 7.59 ± 0.23 | 7.41 ± 0.31 | 7.59 ± 0.23 |
Total virus load: filtrate (log10) | ≤1.75 ± 0.00 | ≤1.75 ± 0.00 | ≤1.75 ± 0.00 |
Conditions | pH 7.0 and 0 M NaCl (low salt) | ||
Minimum reduction value (log10) | ≥5.14 | ≥4.96 | ≥5.11 |
Reduction value (log10) | ≥5.14 ± 0.27 | ≥4.96 ± 0.20 | ≥5.44 ± 0.30≥4.78 ± 0.26 |
Total virus load: load (log10) | 7.35 ± 0.27 | 7.17 ± 0.20 | 7.65 ± 0.306.99 ± 0.26 |
Total virus load: filtrate (log10) | ≤2.21 ± 0.00 | ≤2.21 ± 0.00 | ≤2.21 ± 0.00 |
Conditions | pH 7.0 and 1 M NaCl (high salt) | ||
Minimum reduction value (log10) | 2.74 | 2.77 | 1.15 |
Reduction value (log10) | 2.89 ± 0.30 2.59 ± 0.41 | 2.95 ± 0.35 2.59 ± 0.41 | 1.03 ± 0.41 1.27 ± 0.39 |
Total virus load: load (log10) | 7.41 ± 0.25 | 7.41 ± 0.31 | 7.47 ± 0.29 |
Total virus load: filtrate (log10) | 4.46 ± 0.20 4.76 ± 0.23 | 4.46 ± 0.25 4.82 ± 0.32 | 6.44 ± 0.29 6.20 ± 0.26 |
The LRV difference between high- and low-salt conditions can be traced back to virus adsorption on positive- charged filter media. To understand this adsorptive X-MuLV retention, we calculated a revised LRV with values obtained under high-salt conditions (pH 7.0) subtracted from the value under low-salt conditions (pH 7.0 and pH 5.5) (Table 5, Figure 2). Doing so provided values of 2.2–3.1 LRV for the Zeta Plus depth filters. The Emphaze AEX FNW filter showed revised LRVs that were still high at 4.7 and 4.0 (Table 5, Figure 2). So the main retention mechanism of X-MuLV on the latter at low ionic strength can be attributed to ionic adsorption.
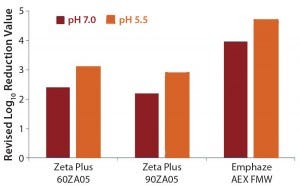
Figure 2: Revised log-reduction values for X-MuLV indicate virus clearance is based on anionic retention mechanisms.
Table 5: Revised log-reduction values for X-MuLV
Filter | Zeta Plus 60ZA05A | Zeta Plus 90ZA05A | Emphaze AEX FNW |
|---|---|---|---|
Conditions | pH 5.5 (clearance attributed to ionic adsorption at low-salt conditions) | ||
Mean value of revised reduction value | 3.10 log10 | 2.89 log10 | 4.69 log10 |
Revised reduction value | 2.95 log10 3.25 log10 | 2.71 log10 3.07 log10 | 4.81 log10 4.57 log10 |
Conditions | pH 7.0 (clearance attributed to ionic adsorption at low-salt conditions) | ||
Mean value of revised reduction value | 2.40 log10 | 2.19 log10 | 3.96 log10 |
Revised (net) reduction value | 2.25 log10 2.55 log10 | 2.01 log102.37 log10 | 3.75 log10 4.17 log10 |
Revised LRVs were lower for Zeta Plus 60ZA05A and 90ZA05A because of the persistent retention (2.7 LRV) at high-salt concentration. That outcome is probably related to hydrophobic interactions and/or marginal mechanical retention. Previous viral clearance studies using Zeta Plus 90ZA filters had already shown that hydrophobic interactions are involved in virus retention at high ionic strengths (6). That phenomenon can be explained by the net charge of X-MuLV, which is nearly zero at pH 7.0 because the heterogenic isoelectric point (pI) of its major capsid protein (p30) ranges from 6.1 to 6.6 (10). Additionally, the p30 protein has many hydrophobic amino acids, possibly presenting exposed hydrophobic moieties.
A minor retention by size may be attributed to convective entrapment that occurs (even if a filter’s pore size is larger than the virus) when viruses are transported to pores by convection but are unable to traverse them fully because of irregular internal structures (11). A further possible reason for LRVs of >1 in the presence of 1M NaCl is related to ionic strength, which is not sufficient to suppress entirely the positive zeta potential. Therefore, some positively charged spots inside the filter might lead to virus removal even at high NaCl concentrations.
With each virus load, two filtration runs were performed in duplicate to provide one value for the load’s total virus load and two values for the filtrates’ total virus load for each tested condition and filter. Reduction values (Equation 1), and a mean value were calculated. The measured total virus load of the filtrates is indicated with mathematical symbols for “equal or less than” (≤) where no virus positive cells were observed with the TCID50 assay sample dilution.
Revised reduction values are calculated by subtracting the means of the reduction values under low-salt conditions (pH 5.5 and pH 7.0) from the means of the reduction values under high-salt conditions (pH 7.0). In the presence of 1M NaCl, electrostatic interactions are widely suppressed independently from the pH value. Therefore, the LRV from the low-salt conditions at pH 5.5 also can be subtracted from the LRV of the high-salt concentration at pH 7.0.
MVM Removal: We observed a very effective MVM removal (4.6–7.2 LRV) for all three depth filters at pH 5.5 and a clearance of 2.4–2.7 LRV at pH 7.0 (Table 6, Figure 3). The nominal pore sizes of all three filters are significantly larger (>0.2 µm) than the MVM virus (20–25 nm), so we believe that such virus-removal capacity is almost solely related to adsorptive retention (3, 12).
Table 6: MVM clearance results for the three depth filters tested
Filter | Zeta Plus 60ZA05A | Zeta Plus 90ZA05A | Emphaze AEX FNW | |
|---|---|---|---|---|
Conditions | pH 5.5 and 0 M NaCl (low salt) | |||
Mean value of reduction value | 7.21 log10 | 6.65 log10 | 4.61 log10 | |
Reduction value (log10) | 6.88 ± 0.29 7.54 ± 0.29 | 6.55 ± 0.32 6.75 ± 0.32 | 4.61 ± 0.34 4.61 ± 0.32 | |
Total virus load: load (log10) | 9.13 ± 0.29 | 8.89 ± 0.32 | 8.83 ± 0.29 | |
Total virus load: filtrate (log10) | 2.25 ± 0.00 1.59 ± 0.00 | 2.34 ± 0.00 2.14 ± 0.00 | 4.28 ± 0.32 4.28 ± 0.26 | |
Conditions | pH 7.0 and 0 M NaCl (low salt) | |||
Mean value of reduction value | 2.56 log10 | 2.74 log10 | 2.42 log10 | |
Reduction value (log10) | 2.56 ± 0.39 2.56 ± 0.38 | 2.74 ± 0.34 | 2.09 ± 0.43 2.74 ± 0.39 | |
Total virus load: load (log10) | 8.65 ± 0.29 | 8.89 ± 0.24 | 8.83 ± 0.29 | |
Total virus load: filtrate (log10) | 6.09 ± 0.26 6.09 ± 0.26 | 6.15 ± 0.24 | 6.74 ± 0.32 6.09 ± 0.26 |
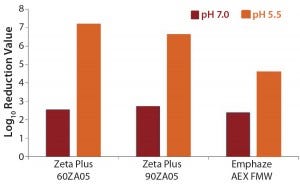
Figure 3: Log-reduction values for MVM at pH 5.5 and pH 7.0
The pI of MVM is about pH 5 (13). So at pH 7.0, the virus should be negatively charged, and at pH 5.5 it should be nearly free of net charge. The higher MVM retention at pH 5.5 indicates that attractive electrostatic effects may play a minor role. It has already been shown that viruses are more effectively removed if operational pH values are near their pI (6).
To explain the mode of virus adsorption in more detail (proportion of ionic and hydrophobic interactions), additional studies can be performed with even higher salt concentrations and/or using organic compounds such as propylene glycol (PG) to suppress electrostatic and hydrophobic interactions. Such studies can improve understanding of the mechanisms responsible for virus retention. However, a detailed breakdown of adsorptive virus removal on a charged depth filter applied in an antibody-purification process that further includes pH virus inactivation and a nanofiltration (size exclusion) is unnecessary to evidence the orthogonality of those removal steps.
With each virus load, our analysts performed two filtration runs in double determination, leading to one value for the load’s total virus load and two values for the filtrates’ total virus load for each tested condition and filter. Reduction values (Equation 1) and mean values were calculated. Measured total virus load of the filtrates is indicated with mathematical symbols for “equal or less than” (≤) where no virus-positive cells were observed in the TCID50 assay sample dilution.
Highly Efficient Virus Removal
We have demonstrated the capability of Zeta Plus 60ZA05A, Zeta Plus 90ZA05A, and Emphaze AEX Hybrid Purifier filters in a unit operation for viral clearance of a MAb purification process. A protein A– purified antibody solution was spiked with two routinely used mammalian model viruses (X-MuLV and MVM), and the downscale virus-retention study was performed under conventional process conditions.
For X-MuLV clearance by Zeta Plus 60ZA05A and 90ZA05A filters, values of 4.7–5.8 LRV could be obtained at pH 5.5 and pH 7.0. We attributed LRVs in the range of 2.2– 3.1 to electrostatic retention mechanisms. The residual virus clearance is probably attributable to hydrophobic or mechanical interactions. The Emphaze AEX FNW filter removed X-MuLV with LRVs of 5.1–5.8 at pH 5.5 and 7.0 (low ionic strength), so its high values of 4.0–4.7 could be attributed to electrostatic retention. The small, nonenveloped MVM virus was reduced efficiently by all tested filters at pH 5.5, with LRVs of 4.6–7.2. MVM clearance was less effective at pH 7.0 (LRV ~2.5).
We found that Zeta Plus and particularly the Emphaze AEX FNW adsorptive depth filters can be considered highly efficient virus- removal options under typical conditions in a MAb downstream process. Moreover, the excellent viral clearance potential of the Emphaze filter by adsorptive retention can be claimed as an orthogonal method for viral safety with the currently adopted virus-filtration step (size exclusion) and low-pH inactivation. The virus removal capability of these charged filters is complementary to other virus clearance steps in downscale validation studies of a MAb process. Just as in validating an anion-exchange chromatography step, the viral- reduction capacity of a charged depth filter must be determined under actual process conditions. So it has to be done individually for specific processes, with different amounts of process-related impurities (HCPs, DNA). We have shown this virus removal capability for our model antibody process using appropriate buffers and conditions.
The capability to cover the purpose of an anion-exchange polishing step with a positively charged depth filter will directly allow us to redesign our three–chromatography-step MAb purification platform into a more efficient and economical two-step process while maintaining product purity, quality, and safety.
Acknowledgments
The authors thank Pia Reimann, Peter Koklitis, and Carsten Erdmann at 3M for their project support and review of this manuscript.
References
1 Zhou JX, et al. Viral Clearance Using Disposable Systems in Monoclonal Antibody Commercial Downstream Processing. Biotechnol. Bioeng. 100(3) 2008: 488–496.
2 ICH Q5A (R1). Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. US Fed. Reg. 63(185) 1998: 51074.
3 Bergmann K, et al. Design and Performance of Viral Clearance Studies. BioProcess Int. 4(10) 2006: 56–62.
4 Faude A, et al. Virus Removal By Simple Depth Filtration in a Two-Step Downstream Process for MAb Production (poster). Recovery Conference, Rostock, 2014; http://rentschler.de/fileadmin/Downloads/ Rentschler_Poster_Recovery_201407.pdf.
5 Wang M. Zeta Plus™ VR Filters for Viral Reduction. BioProcess Int. 9(7) 2011: 62.
6 Zhou JX. Orthogonal Virus Clearance Applications in Monoclonal Antibody Production. Process Scale Purification of Antibodies. Gottschalk U, Ed. John Wiley & Sons: Hoboken, NJ, 2009; 169–186.
7 Revie D, et al. Novel Application Approaches to Obtain Maximum Viral Clearance from an Immunoglobulin Production Process. IBC’s Second International Symposium on Viral Clearance, 1998.
8 Application Brief. Cuno Zeta Plus®VR Filters for Viral Reduction in Blood Plasma Fractionation Processes. Cuno (3M): Meriden, CT, April 2002; http://multimedia.3m.com/ mws/media/418846O/zeta-plus-vr-filters-viral- reduction-blood-plasma-fractionation. pdf?fn=DOC00042%20-%20 LITCABZPVR1.E.pdf.
9 Müller D, et al. Taking MAb Purification to the Next Level. Europ. Biotechnol. Life Sci. Ind. 13, 2014: 76.
10 Mitra S, et al. Comparative Physicochemical and Biological Properties of Two Strains of Kilham Rat Virus, a Non-Defective Parvovirus. J. Gen. Virol. 61(Pt l) 1982: 43–54.
11 Trilisky, et al. Flow-Dependent Entrapment of Large Bioparticles in Porous Process Media. Biotechnol. Bioeng. 104(1) 2009: 127–133.
12 Hou K, et al. Capture of Latex Beads, Bacteria, Endotoxin, and Viruses By Charge- Modified Filters. Appl. Environ. Microbiol. 40(5) 1980: 892–896.
13 Ray S, et al. Chapter 26: Single-Use Virus Clearance Technologies in Biopharmaceutical Manufacturing: Case Studies. Single-Use Technology in Biopharmaceutical Manufacture. Eibl R, Ed. John Wiley & Sons: Hoboken, NJ, 2011: 311–322.
Mario Metzger and Anja Gerster are process engineers in technology development; Marcus Peiker is process manager, Sabine Faust is a process engineer, and Alexander Faude is associate director of downstream process development; Sybille Ebert is assicate director, and corresponding author Dr. Dethardt Müller (dethardt. [email protected]) is vice president of technology development at Rentschler Biotechnologie GmbH in Laupheim, Germany. Dr. Sophie Winterfeld ([email protected]) is senior engineer in application development for life sciences process technologies, and Nicole Mang is a senior sales account executive in life-science process technologies at 3M Deutschland GmbH in Neuss, Germany.
You May Also Like





