The Promise of Artificial Intelligence in HealthcareThe Promise of Artificial Intelligence in Healthcare
 The term artificial intelligence (AI) has become pervasive in conversations about the future of healthcare. AI has the potential to transform medicine through novel models of scientific discovery and healthcare delivery, ultimately leading to improved individual and public health. Yet misunderstanding and miscommunication abound. Thus, concepts related to AI need to be defined and explained to elevate our general level of understanding and our discourse around the topic.
The term artificial intelligence (AI) has become pervasive in conversations about the future of healthcare. AI has the potential to transform medicine through novel models of scientific discovery and healthcare delivery, ultimately leading to improved individual and public health. Yet misunderstanding and miscommunication abound. Thus, concepts related to AI need to be defined and explained to elevate our general level of understanding and our discourse around the topic.
The Promise of AI in Healthcare
AI has been studied by computer scientists for over 70 years. The term itself was coined by John McCarthy in 1956 at the first workshop on the subject at Dartmouth College (1). But the theory and topics that became known as AI have a much longer history (2). Even so, it remains one of the most complex and misunderstood topics in computer science because of the vast number of techniques used and the often-nebulous goals being pursued.
AI and healthcare have been bound together for over half a century. The Dendral project was an early expert system based on AI techniques from Stanford in the 1960s. Its was used for scientific hypothesis formation and discovery. The primary focus was to determine organic-compound structures by analyzing their mass spectra (3). Dendral was followed by Mycin in the 1970s (also by Stanford University) with the goal of identifying infection-causing bacteria and to recommend antibiotics, with dosage adjusted to patient weights. The concepts behind Mycin were generalized to all internal medicine in the 1980s with the Caduseus system (developed by Harry Pople at the University of Pittsburgh). At the time, it was described as the “most knowledge-intensive expert system in existence” (4). In parallel, techniques based on such programs were developed for drug discovery.
Since those early programs, techniques and healthcare-use cases have developed against a wider backdrop of AI “summers and winters” — an apt metaphor for the cyclical interest in AI measured against expectations or hype and the realities of AI implementation. Currently, some industries, including bioprocessing, are experiencing an unprecedented AI summer that many believe is an integral part of the digital age, often called the Fourth Industrial Revolution. For companies that develop and manufacture biopharmaceuticals, it is known as Bioprocessing 4.0.
All major healthcare and life science organizations have used or are investigating AI-enabled applications. The current success (and hype) of AI is driven largely by increases in computing power, availability of cheap storage and fast networking, advancement of algorithms, increasing data capture mechanisms, and increased awareness among consumers. However, navigating the growing interest in such a large subject demands a well-defined set of foundational concepts and terms.
AI has made the most advances in applications with access to substantial structured data and in cases where the problem to solve is well understood or straightforward to define (e.g., image recognition and language translation). The opposite is true for most cases in healthcare, where data are hard to obtain because they are expensive, restricted, and often incomplete or fractured among different stakeholders. Typically, life-science data are complex, inherently high-dimensional, and semistructured or unstructured. In such cases, the questions to answer are not simple to frame.
Nonetheless, the potential of AI to influence patient quality of life dramatically continues to drive the development, validation, and implementation of AI in healthcare settings. Implementation will require detailed attention to safety and efficacy, so broad adoption will take time and effort to get right. Below, we hope to serve that goal by orienting newcomers to the current understanding and development efforts for applications of AI in healthcare.
The first step in elevating understanding is to settle on definitions. Merriam-Webster defines artificial intelligence as “the capability of a machine to imitate intelligent human behavior” (5). That definition is fraught with issues, most of all being the comparison with “human behavior,” which is itself ill defined. Further, the brief definition limits the concept of intelligence, which is not exclusively human. Currently, contrived agents perform tasks in supplement to and often above “expert” human counterparts (6). In his book, Max Tegmark refers to AI in the abstract as anything that is “nonbiological [and has the] ability to accomplish complex goals” (7). That concise definition also is problematic because a great deal of recent work, both theoretical and practical, has been performed on biologically based “intelligent agents” such as a DNA-based agent (8).
For such reasons, we at the Alliance for Artificial Intelligence in Healthcare (AAIH) prefer the more mundane (but clear cut) definition of AI as “the study of artificial intelligent agents and systems, exhibiting the ability to accomplish complex goals.” All machine learning (ML) systems consist of training data, learning algorithms, and a resulting model representation.
ML models can take in and train on more data than a person can. ML models can operate at speeds and scales well beyond human capability. ML algorithms can build far more complex models than a human could. ML is data-driven, and the models can be applied to all narrow problems given proper training data. Because of the complexity of biology, the rate at which new knowledge is being generated in healthcare, and the reaction speed needed in time-critical decision making, ML is a tool that can help scientists, clinicians, and medical professionals along the spectrum of biomedical discovery, clinical development, patient care, and population health to make good decisions.
ML helps biomanufacturers reduce failure rates and lower drug development costs by increasing the number and quality of available targets, designing and testing molecules that are effective at treating disease with minimal toxicity or adverse events, and selecting the right patients at the right times for the right treatments in clinical trials. In clinical care, ML drives efficiencies in clinical workflows. It also plays a significant role in aiding decision making among health practitioners along the continuum of prevention, diagnosis, treatment, and patient follow-up. ML permits early, accurate diagnosis by aggregating disparate pieces of information and extracting key patterns from data sets to help identify effective interventions early on — when conditions are amenable to treatment.
To produce an ML model, a representation of your data must be selected. When possible, each parameter in that representation should relate to the underlying phenomenon being modeled in a meaningful way. For example, a model of cancer risk would have a set of meaningful parameters including age, tobacco consumption, weight, alcohol consumption, and so on. Examples of parameters unrelated to cancer risk would be color of car or favorite music style. Inclusion of parameters unrelated to a phenomenon can be problematic because ML algorithms look for correlations. An unrelated parameter is statistically likely to contain a spurious correlation given that training sets are finite in size. That can lead to biases and false conclusions.
To illustrate potential applications and underlying decisions that drive AI adoption, the series of case studies below focus on how AI has and can be used to advance healthcare. This sampling represents applications in drug development.
Pharmacovigilance
Pharmacovigilance (PV) is the study of adverse effects from pharmaceutical products after dosage administration. Almost all markets for pharmaceutical products have regulations on collecting and studying PV data, both pre- and postapproval. With the increase in human longevity, improved access to pharmaceuticals, and emergence of internet-connected monitoring devices, the amount of PV data being processed by companies related to their products is increasing rapidly.
Approach: Pfizer concluded that case-processing activities (extracting data using natural-language processing from submitted documents) constitute up to two-thirds of internal PV resources. To improve the cost and efficiency of case processing, the company compared PV AI solutions from three vendors with that from its internal AI center of excellence (9).
Solution: In a pilot system, the vendors’ AI systems were trained using a set of 50,000 correctly annotated documents. Then the systems were tested on 5,000 unannotated test documents and compared with the hand-processed annotated version. In the second cycle, the amount of training data was doubled to establish whether the systems would become more accurate as additional training data were used (Figure 1).
Business Results: The study found that two vendors had an accuracy rate >70% in extracting information from PV data test sets. Case-level accuracy showed that the same two vendors processed 30% of cases with >80% accuracy and showed improvement in their second cycle.
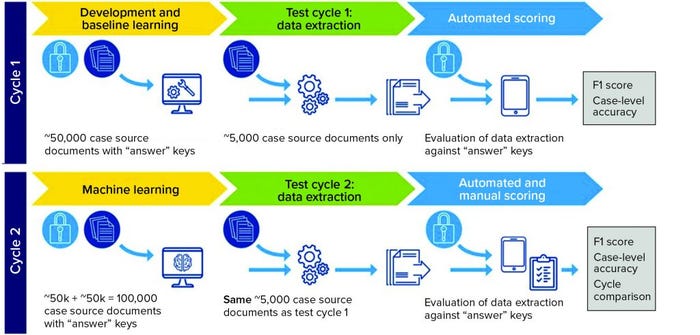
Figure 1: Pilot system set up (F1 score is the harmonic mean of precision and recall, taking into account both false positives and false negatives.)
Forecasting Cell Culture
Cell culture is an important step in biomanufacturing therapeutic antibodies, metabolites, and other biologics. Chinese hamster ovary (CHO) cell lines are used to express recombinant proteins for a number of drug products, including vaccines and anti-PD-L1 checkpoint inhibitor antibodies (pembrolizumab). CHO cells and most expression platforms are subject to inherent variability caused by biological and abiotic factors. That can cause inconsistencies in production quality and quantity, potentially leading to costly shutdowns or batch failures.
Approach: ML models trained on historical production data (e.g., process conditions and real-time monitoring data) can be used to forecast production outcomes and catch detrimental variations before they cascade into downstream failures.
Solution: Schmitt and colleagues (from bioprocessing giants Lonza and Pfizer) devised a classification model that could predict whether a run would result in lactate consumption or accumulation (10). While building their model, the researchers also identified novel hypotheses into the metabolic mechanisms of lactate production. The result of this modeling effort was a dynamic model predictive controller (MPC) to modulate lactate production (Figure 2).
Business Results: The ML-derived MPC enabled bioprocessing engineers to control lactate accumulation in bioreactors, thereby improving specific productivity of a cell culture. Predictive models coupled with dynamic controls could benefit biomanufacturing processes, enabling optimization of production systems, decreasing failure risk, and facilitating error- free automation. Such capabilities could provide economic benefits. With the biomanufacturing industry’s need to scale up production of many drug products (e.g., vaccines), such ML-enabled scaling can benefit public health as well.
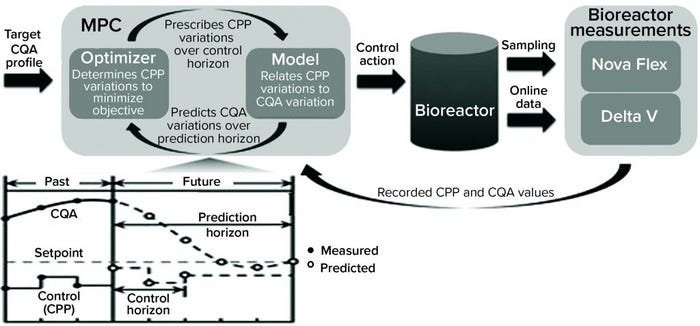
Figure 2: Bioreactor-integrated model predictive controller (MPC) (10) (CPP = critical process parameter, CQA = critical quality attribute; Nova Flex and Delta V are proprietary systems of Nova Biomedical and Thermo Fisher Scientific, respectively.)
Small-Molecule Generation
Small-molecule drug discovery requires a vast search space and a large number of design–make–test cycles to find a single candidate molecule. Candidates must meet different stringent criteria for in vitro and in vivo characteristics and behavior. Producing, or “inventing,” molecules with optimal drug-like properties instead of searching for one is exactly the idea being pursued by a number of groups using generative adversarial networks (GANs).
The first peer-reviewed publications on the application of GANs to generative chemistry used molecular fingerprint representation and an adversarial autoencoder (AAE) architecture (11, 12) as well as string-based representations using variational autoencoders (VAE) (13, 14). Later versions combined GANs and reinforcement learning by introducing the objective reinforced generative adversarial network (ORGAN) architecture for generating novel molecules (15). The approach was extended by introducing adversarial threshold (AT) (16) and differential neural computer (DNC) (17) concepts. GANs can generate novel molecules using a graph representation of a molecular structure (18) and three-dimensional representation (19).
Approach: Zhavoronkov et al. developed a generative tensorial reinforcement learning (GENTRL) neural network for de novo lead-like molecules design with high potency against a protein of interest (Figure 3) (20). They discovered potent inhibitors of discoidin domain receptor 1 (DDR1) (a kinase target implicated in fibrosis and other diseases) within 21 days.
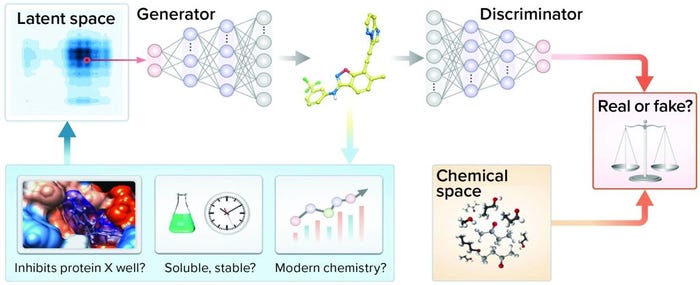
Figure 3: Representation of the generative adversarial networks (GANs) and reinforcement learning (RL)
Solution: Reinforcement learning, variational inference, and tensor decompositions approaches were combined into a generative two-step ML algorithm named GENTRL. The first step was to train an autoencoder-based model to learn a mapping from a discrete molecular space to a continuous parameterized space. At that stage, the network was trained on the general medicinal chemistry data set together with kinase chemistry and patent data. Medicinal chemistry evolution 2018 (MCE-18), half maximal inhibitory concentration (IC50), and medicinal chemistry filters were used to parameterize learned representation (21).
At the second stage, the model was fine-tuned to expand the latent manifold toward discovering novel inhibitors and preferentially generate DDR1 kinase inhibitors. Reinforcement learning was applied with reward functions based on self-organizing maps (SOM).
The trained GENTRL model generated 30,000 novel unique molecular structures, which were filtered with the prioritization pipeline down to 40 molecules selected for synthesis and real-world experiments (Figure 4). Out of six remaining molecules, two had two-digit nanomolar IC50 and high specificity against DDR1 protein.
Business Results: The generative network now can be used to generate compounds with desired physicochemical properties, thus reducing the need for manually generating combinatorial libraries of compounds. The strategy could increase the efficiency of lead generation by one or more orders of magnitude and help researchers identify compounds to test against previously hard-to-drug targets across different disease areas.
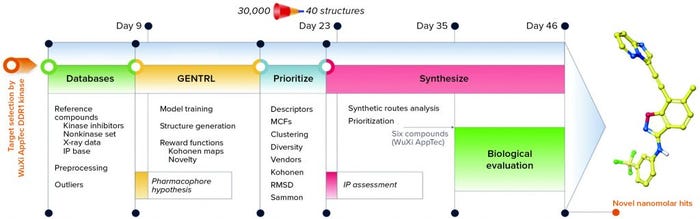
Figure 4: Workflow and timeline of the discoidin domain receptor 1 (DDR1) project (GENTRL = generative tensorial reinforcement learning, IP = intellectual property, MCF = medicinal chemistry filters, RMSD = root-mean-square deviation)
Modeling Side Effects of Drug Combinations (Polypharmacy)
Some diseases and medical conditions are treated with drug combinations. For treatments against cancer, for example, combination therapies are being explored to find targeted therapies and biologics to pair with chemotherapy or with one another. Some drugs such as checkpoint inhibitors (CPIs) (e.g., pembrolizumab) have attained blockbuster success yet fail to improve outcomes for most patients. Thus, an entire cottage industry has grown to search for the right drugs to combine with CPIs to expand their effectiveness in patients. Another example is the common scenario in which a patient takes multiple drugs as part of their daily regimen, and polypharmacy increases the chance of deleterious side effects because of drug–drug interactions.
Adverse events are a major concern in drug development. Every drug must pass stringent toxicity criteria before approval. Yet testing all possible drug pairs for adverse events induced by drug–drug interactions is impractical, and many clinical trials are too small to detect rare but serious polypharmacy effects. Over 15% of the US population is influenced by polypharmacy. Treating the unexpected consequences costs hundreds of billions of US dollars.
Approach: Zitnik and colleagues at Stanford (21) developed a graph convolutional network (GCN) approach to model drug–drug interactions and to predict specific side effects and complications associated with such interactions. Their model uses information from drug–protein (target) and protein–protein interactions. Those relationships were meaningful in the prevalence of multidrug prescriptions. A key innovation of the model was that it not only predicted whether to expect a drug–drug interaction or polypharmacy side effect, but also identified the specific type of interaction to expect from among nearly 1,000 defined adverse side effects.
Solution: The authors presented Decagon, a solution to a “multirelational link prediction problem in a two-layer multimodal graph/network of two node types: drugs and proteins” (Figure 5). Data sets were assembled from published sources; different protein–protein interaction networks; STITCH database for drug–protein interactions; and Sider, Offsides, and Twosides databases for drug–drug interactions. Once the authors harmonized vocabularies, the resulting network included 645 drug and 19,085 protein nodes linked by 715,612 protein–protein, 18,596 drug–protein and 4,651,131 drug–drug edges.
The Decagon model consists of an encoder and decoder. The encoder is a GCN that operates on the graph of protein–drug nodes and edges that create embeddings for the nodes. The decoder uses tensor factorization to translate the node embeddings into edges to predict the likelihood and type of polypharmacy events (drug–drug interactions). During training, model parameters are optimized by cross-entropy loss, with 80% of the data used for training. The rest is removed for parameter selection and testing or validation. The Decagon network outperformed other tensor-factorization and neural-embedding approaches to polypharmacy prediction by a wide margin. Its average area-under-the-receiver operating characteristic (AUROC) score across all 964 side-effect types was 0.872, with the nearest comparator at 0.793. The model performed especially well for side effects with strong molecular bases.
Business Results: The Decagon model could be used in primary care settings as a decision-support tool for physicians to guide prescriptions for patients taking several drugs. Alternatively, the model could be helpful in designing drug-combination clinical trials. Eventually, a model of this type might also serve a regulatory function, such as for contraindication labeling.
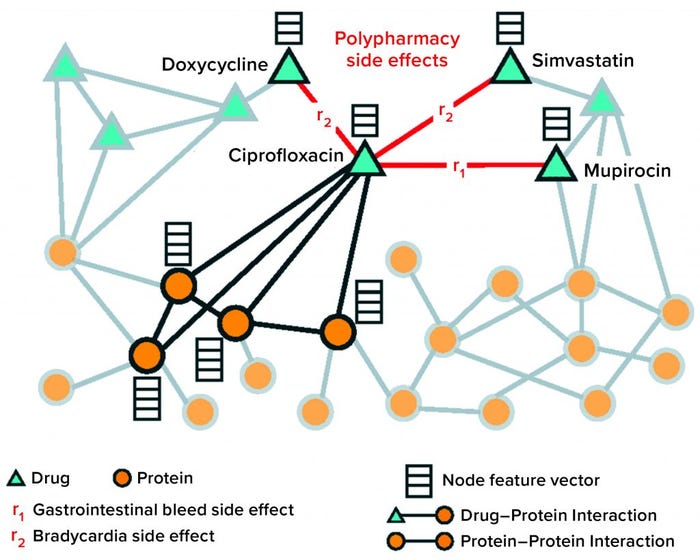
Figure 5: Nodes and edges comprising protein and drug graph operated on by the Decagon graph convolutional neural network
The Future of AI
AI tools have been applied across different healthcare use cases, including aiding in drug discovery, improving clinical outcomes, and optimizing biomanufacturing. In the coming years, we expect the bioprocess industry to face increasing competitive pressure to leverage AI for optimizing and shortening product development times. The long and bumpy road from preclinical discovery to market ideally will be shortened and smoothed by intelligent agents helping with design, discovery, optimization, positioning, and production. Downstream failures will be predicted before a product ever enters a laboratory, clinic, or bioreactor. The AI-enabled future not only will disrupt traditional approaches to biomedical discovery and healthcare delivery, it will accelerate progress in cutting-edge fields such as synthetic biology and metabolic engineering, cell and gene therapies, and novel biologics.
With advances in protein-structure calculation courtesy of Deepmind’s Alphafold (23), biomanufacturers will begin to observe a heavy emphasis on computing biologic functions and toxicity — not only from a structural standpoint, but also from a systems biology perspective. Solving the challenges of systems biology will require a massive number of calculations, either from supercomputers or the oncoming quantum computing revolution. Far more options in biologics formulation will be available because the successes and failures of the past two decades can be used to teach computers to optimize excipient selection and offer a wide range of choices for drug delivery.
Similarly, biomanufacturing will be transformed by combining biologic sequence data with cell-growth kinetics to predict the precise conditions and reagents that will maximize yield. And clinical trial patient selection will become more precise, moving the industry away from attempting to discover panaceas and toward personalized medicines.
Given the potential benefits of AI in healthcare, but also its real possibility to cause harm, we call for a concerted and collaborative effort to improve industrywide understanding of the complexities of AI. Critically, the healthcare industry should work together with governments and patients to advance the discussion of responsible AI use. We must work to develop standards that will ensure trustworthiness and transparency in decisions supported by AI as we promote the use of AI in all aspects of healthcare to ensure the best possible decisions always are made.
References
1 Moor J. The Dartmouth College Artificial Intelligence Conference: The Next Fifty Years. AI Magazine 27(4) 2006: 87–91; https://ojs.aaai.org//index.php/aimagazine/article/view/1911.
2 Nilsson NJ. The Quest for Artificial Intelligence. Cambridge University Press: Cambridge, UK, 2010; https://doi.org/10.1017/CBO9780511819346.
3 Lindsay RK, et al. Dendral: A Case Study of the First Expert System for Scientific Hypothesis Formation. Artificial Intelligence 61(2) 1993: 209–261; https://doi.org/10.1016/0004-3702(93)90068-M.
4 Feigenbaum EA, McCorduck P. The Fifth Generation: Artificial Intelligence and Japan’s Computer Challenge to the World. Addison-Wesley: Boston, MA, 1983.
5 Artificial Intelligence. Merriam-Webster Dictionary. Merriam-Webster: Springfield, MA; https://www.merriam-webster.com/dictionary/artificial%20intelligence.
6 Goertzel B. Who Coined the Term “AGI”? August 2011; https://web.archive.org/web/20181228083048/http://goertzel.org/who-coined-the-term-agi.
7 Tegmark M. Life 3.0: Being Human in the Age of Artificial Intelligence. Knopf Doubleday Publishing Group: New York, NY, 2017.
8 Qian L, et al. Neural Network Computation with DNA Strand Displacement Cascades. Nature 475, 2011: 368–372; https://doi.org/10.1038/nature10262.
9 Schmider J, et al. Innovation in Pharmacovigilance: Use of Artificial Intelligence in Adverse Event Case Processing. Clin. Pharm. Thera. 105(4) 2019; https://doi.org/10.1002/cpt.1255.
10 Schmitt J, et al. Forecasting and Control of Lactate Bifurcation in Chinese Hamster Ovary Cell Culture Processes. Biotechnol. Bioeng. 116(9) 2019: 2223–2235; https://doi.org/10.1002/bit.27015.
11 Kadurin A, et al. The Cornucopia of Meaningful Leads: Applying Deep Adversarial Autoencoders for New Molecule Development in Oncology. Oncotarget 8(7) 2016: 3098–3104; https://doi.org/10.18632/oncotarget.14073.
12 Kadurin A, et al. druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico. Mol. Pharm. 14(9) 2017: 10883–10890; https://doi.org/10.1021/acs.molpharmaceut.7b00346.
13 Gomez-Bombarelli R, et al. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 4(2) 2018: 268–276; https://doi.org/10.1021/acscentsci.7b00572.
14 Lim J, et al. Molecular Generative Model Based on Conditional Variational Autoencoder for de Novo Molecular Design. J. Cheminform. 10(1) 2018: 31; https://doi.org/10.1186/s13321-018-0286-7.
15 Benjamin SL, et al. Optimizing Distributions over Molecular Space: An Objective-Reinforced Generative Adversarial Network for Inverse-Design Chemistry (ORGANIC). ChemRxiv August 2017; https://doi.org/10.26434/chemrxiv.5309668.v3.
16 Putin E, et al. Adversarial Threshold Neural Computer for Molecular de Novo Design. Mol. Pharm. 15(10) 2018: 4386–4397; https://doi.org/10.1021/acs.molpharmaceut.7b01137.
17 Putin E, et al. Reinforced Adversarial Neural Computer for de Novo Molecular Design. J. Chem. Inf. Model 58(6) 2018: 1194–1204; https://doi.org/10.1021/acs.jcim.7b00690.
18 De Cao N, Kipf T. MolGAN: An Implicit Generative Model for Small Molecular Graphs. Cornell University, 2018; https://arxiv.org/abs/1805.11973.
19 Kuzminykh D, et al. 3D Molecular Representations Based on the Wave Transform for Convolutional Neural Networks. Mol. Pharm. 15 (10), 2018; 4378–4385; https://doi.org/10.1021/acs.molpharmaceut.7b01134.
20 Zhavoronkov A, et al. Deep Learning Enables Rapid Identification of Potent DDR1 Kinase Inhibitors. Nat. Biotechnol. 37(9) 2019; 1038–1040; https://doi.org/10.1038/s41587-019-0224-x.
21 Ivanenkov Y, et al. Are We Opening the Door to a New Era of Medicinal Chemistry or Being Collapsed to a Chemical Singularity? J. Med. Chem. 62(22) 2019: 10026–10043; https://pubs.acs.org/doi/full/10.1021/acs.jmedchem.9b00004.
22 Zitnik M, Agrawal A, Leskovec J. Modeling Polypharmacy Side Effects with Graph Convolutional Networks. Bioinformatics 34(13) 2018: i457–i466; https://doi.org/10.1093/bioinformatics/bty294.
23 AlphaFold: A Solution to a 50-Year-Old Grand Challenge in Biology. DeepMind 30 November 2020; https://deepmind.com/blog/article/alphafold-a-solution-to-a-50-year-old-grand-challenge-in-biology.
Corresponding author Oscar Rodriguez is vice president of Data Engineering at BlackThorn Therapeutics; [email protected]; https://www.blackthornrx.com. Rafael Rosengarten, PhD, is CEO of Genialis, [email protected]; https://www.genialis.com. Andrew Satz is the CEO of EVQLV; [email protected]; https://evqlv.com.
This article is an abridged version of “Artificial Intelligence in Healthcare: A Technical Introduction,” written by Brandon Allgood, Oscar Rodriguez, Jeroen Bédorf, Pierre-Alexandre Fournier, Artur Kadurin, Alex Zhavoronkov, Stephen MacKinnon, Rafael Rosengarten, Michael Kremliovsky, Aaron Chang, and Annastasiah Mudiwa Mhaka. New contributions were provided by Andrew Satz.
You May Also Like





