Designing Vaccines: The Role of Artificial Intelligence and Digital Health, Part 1Designing Vaccines: The Role of Artificial Intelligence and Digital Health, Part 1
 According to the founder and executive chairman of the World Economic Forum, Klaus Schwab, the fourth industrial revolution began in the 21st century and is characterized by an unprecedented development and exponential growth of a high-technology industry transforming society at every level (1–4). In particular, healthcare systems are evolving rapidly to adapt to the new reality. According to Forbes, the main technologies currently shifting the paradigm of medical research are artificial intelligence (AI) and machine learning (ML) (5), both defined in the “Terms and Abbreviations” box. From a marketing perspective, experts predict that the global AI healthcare market will grow from US$4.9 billion in 2020 to reach $45.2 billion by 2026 (6).
According to the founder and executive chairman of the World Economic Forum, Klaus Schwab, the fourth industrial revolution began in the 21st century and is characterized by an unprecedented development and exponential growth of a high-technology industry transforming society at every level (1–4). In particular, healthcare systems are evolving rapidly to adapt to the new reality. According to Forbes, the main technologies currently shifting the paradigm of medical research are artificial intelligence (AI) and machine learning (ML) (5), both defined in the “Terms and Abbreviations” box. From a marketing perspective, experts predict that the global AI healthcare market will grow from US$4.9 billion in 2020 to reach $45.2 billion by 2026 (6).
AI makes it possible to turn the challenge of big-data growth within the biomedical sector into an advantage. It is not surprising that application of AI already has been integrated into drug-discovery processes: 40% of drug-discovery start-ups already are exploiting AI to identify new drug candidates, 28% use it to identify new targets, and 17% use it for de novo drug design (7). Moreover, the first drug solely designed by AI — DSP-1181 — has entered the phase 1 clinical trial stage. According to its developer, Exscientia, using AI enabled the drug’s discovery in a record 12 months; a conventional process would take four to six years (7).
AI is integrating and changing drug development at a fast pace justified not only by unmet medical needs. Pharmaceutical companies are integrating advanced technologies and investing into highly lucrative development programs, whereas vaccine development programs of importance to global health remain severely underserved (8).
Vaccines have changed human history and currently save two to three million human lives per year, according to the World Health Organization (WHO) (9). R. Rappuoli achieved one milestone in vaccine engineering in 1990 by exploiting advances in sequencing technology — using reverse vaccinology (RV) — to identify surface-expressed vaccine candidate antigens of meningitis B (MenB). The RV method has transformed vaccine development and last year brought marketing authorization (MA) for the first RV-developed MenB vaccine (10).
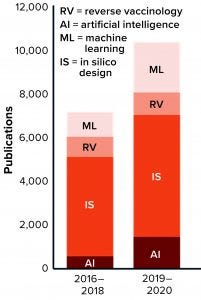
Figure 1: The number of publications that incorporate terms related to artificial intelligence (AI) for vaccine development is growing.
The number of publications in vaccine development mentioning “AI,” “ML,” “RV,” and “in silico vaccine” has grown in the past five years, indicating that such technologies are being integrated slowly into vaccine development programs (Figure 1). Despite all progress, however, many “old” vaccine-specific challenges remain unresolved while new, unforeseen obstacles arise to imperil global health and the worldwide economy. Some infectious diseases remain unpreventable by inoculation (e.g., human immunodeficiency virus, HIV). Some current vaccines need significant improvement in efficacy (e.g., bacille Calmette-Guérin, BCG, for tuberculosis). New vaccines are needed against chronic infections (e.g., herpes simplex virus, HSV). And finally, the world will need vaccines against emerging and reemerging infectious diseases (e.g., Chikungunya, Dengue, Zika, and potential pandemics).
Vaccine development is a well-recognized, low-margin market that requires paramount investment into product development and approval. Essentially, the establishment and maintenance of new vaccine pipelines mostly relies on industry sponsorship. But the development costs from basic research to MA can be $200–500 million, not including additional costs of failures and postapproval regulatory demands for additional studies (11).
Therefore, all vaccine manufacturers struggle to make decisions regarding which vaccine candidates to develop next. The risk of failure is tremendous, so any tool that would facilitate such decisions would be highly beneficial. This review focuses on ways to leverage AI power for saving time and resources in vaccine development.
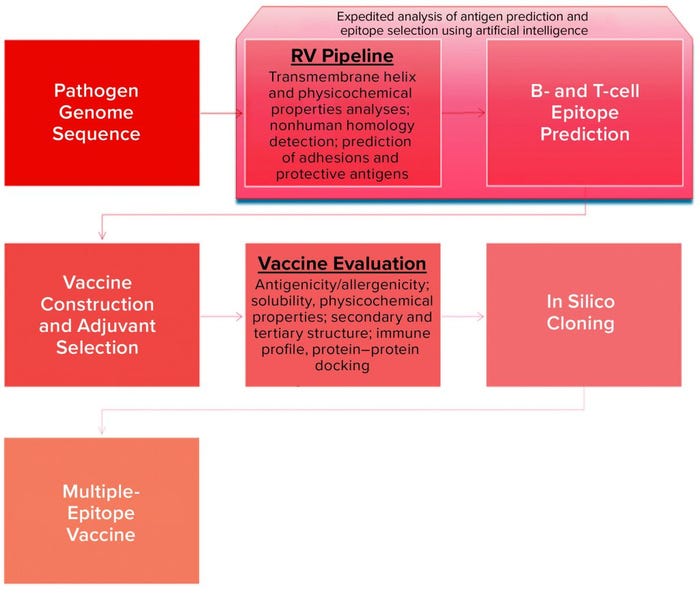
Figure 2: Schematic view of in silico platform for vaccine design (26, 31)
Current Challenges and AI Implications
Discovery Phase: The landscape of vaccine development has been transformed through advances in DNA recombination as well as computational and structural biology (e.g., reverse and structural vaccinology). A number of computing tools can address the increasing demand to process the large volume of complex biological data generated by the scientific community in both translational and clinical research (12–14). Today, virtual technological tools and AI power can be used to design vaccine candidates without ever entering a “wet” laboratory (Figure 2), even including in silico trials (15). Widely available pathogen genome sequences provide the basis for selection of appropriate protein antigens through proteome analysis in RV approaches. Covering the physiochemical properties of protein sequences, VaxiJen software — the first suite of tools (or platform) to exploit ML strategy — enables prediction of suitable bacterial and viral antigens (16, 17). Although RV has been successful for a MenB vaccine, however, a recent comparison analysis of available RV software platforms revealed poor accuracy to match that predictive success with known bacterial protective antigens (BPAs) (18).
Critical steps in vaccine design include the selection of antigens that correspond optimally with our innate (pattern recognition receptor epitopes) and adaptive (the B- and T-cell epitopes) immune systems to incur a productive and long-lasting immune response. Epitope mapping has to be performed for a lead candidate, but the typical experimental techniques applied for that purpose — crystallography, mass spectrometry (MS), nuclear magnetic resonance (NMR), and antibody-based methods — are costly and time-consuming (19). Antigen–antibody (Ag–Ab) in silico screening using computational techniques offers a superior high-throughput alternative. AI and ML algorithms can build three-dimensional (3D) models representing the interactions between antigens and major histocompatibility complex (MHC) molecules (20). EpiXax (iVAX), IEDB, SYFPEITHI, JanusMatrix, and other in silico platforms have been developed to assist with epitope mapping and immunogenicity prediction (21, 22). You can find an extensive review of available software for vaccine development elsewhere (23–25), so the options will not be detailed herein.
AI could speed up and facilitate in silico vaccine design by providing a shortcut through merging several steps into a one. As depicted in Figure 2, that can lead to extremely accurate and efficient prediction of vaccine candidates. Recent approaches include the DeepVacPred vaccine-design platform based on a deep neural-network architecture that uses virus protein sequences to design promising vaccine candidates within a second (26).
Adjuvants can enhance vaccine potency greatly. As demonstrated recently, ML enables individualized prediction of an immune response for different options to facilitate adjuvant selection (27). Essentially, computational biology immunoprofiling can help identify fine-tuned differences in adjuvant-mediated immune response that can be overlooked using conventional readouts — thus paving the way toward discovery of new biomarkers to detect functional postimmunization immune responses (28).
Rapid evaluation of vaccine candidates based on reliable prediction of reactogenicity and immunogenicity could offer benefits in time and cost efficiencies from the magnitude of ML techniques available (29). Currently, only limited integration of mathematical modeling in the initial stages of vaccine development is possible. But it could help to shape vaccine-development processes within a tight and goal-oriented frame, ultimately minimizing the risks of failure. Mathematical modeling could help to define key parameters describing desired health impacts for a given medicinal product, thus guiding the design of target product profiles (TPPs) (30).
Novel development programs for vaccines increasingly are integrating contemporary strategies such as immunoinformatics, comparative proteomics, in silico hierarchical approaches, RV, system vaccinology, and mathematical modeling (14). But it is important to note that all those strategies have been applied (or at least proposed) only in the initial design stage or have reached only the very early phase of preclinical testing. Therefore, no data are available yet to estimate the real contribution of innovative designs toward success of final products.
Preclinical Testing: The preclinical phase of vaccine development is intended to test safety, immunogenicity, and efficacy of lead vaccine candidates in relevant animal models before the first-in-human clinical phase. The traditional approach focuses on proof of concept and lead-candidate optimization, with evaluations of toxicity, immunogenicity, and efficacy relying on the availability of suitable animal models (typically mice, rabbits, and nonhuman primates) (32). Over the past decade, animal testing has been reduced significantly thanks to joint efforts of regulatory agencies to encourage the use of animal-free “new-approach methods” (NAMs). Such alternative models use advanced in vitro modeling techniques — essentially, computational approaches. Their application and development serve not only to address ethical considerations based on the “3R” principle (replacement, reduction, refinement), but also to accelerate the speed of preclinical testing (33). Moreover, when therapeutic demand is sudden and great (e.g., during a pandemic), available time is too short to perform lengthy and exhaustive nonclinical investigations.
Considerable progress has been documented in shifting the toxicity-testing paradigm of chemical compounds in the United States (2007 National Research Council Report) and European Union (European Centre for the Validation of Alternative Methods) (34, 35). Substantial replacement of animal toxicology tests could be achieved through prediction of compound toxicities based on ML algorithms (36). Combining in vitro risk-assessment techniques with systems biology could facilitate predictions of drug safety (37, 38). Although small molecules and vaccines follow similar preclinical testing algorithms, some drawbacks are arising in preclinical vaccine evaluation. Those drawbacks are associated with the preventive modalities inherent to vaccines, which function through engagement of a host’s immune system. Immunogenicity testing as part of toxicology evaluations becomes a major focus of vaccine candidate investigation.
Vaccine-specific parameters that should be addressed primarily in the preclinical phase include characterization of a vaccine-induced immune response, vaccine efficacy, and product safety. A functional immune system becomes prerequisite to addressing those critical product attributes, thus limiting them to in vivo testing. With the immense complexity of interactions between antigens and our immune systems, replacing in vivo validation with virtual means appears to be unrealistic (39). Nevertheless, system approaches could improve some aspects significantly. Here are just a few examples:
assisting in the choice of suitable animal models to mimic closely and/or extrapolate the progress of disease in humans
applying to planning of studies that reach sufficient statistical power (small or large animal species)
identifying routes of pathogen exposure and administration, and helping to set dosages
defining the most predictive clinical end point
dissecting correlates of protection (which are not identified for most vaccines) and identifying new biomarkers that predict vaccine efficacy
anticipating immunogenicity in humans based on animal data
determining vaccine efficacy based on titers of neutralizing antibodies in cases of acknowledged correlation (e.g., hepatitis A and B)
facilitating the iterative design of preclinical studies based on identification of parameters (e.g., microbiomes, comorbidities, and genetic diversity) relevant to vaccine-induced immune response and based on metadata analysis (using AI) to improve databases derived from animal models.
Overall, computational technologies and AI could improve preclinical study designs greatly through extraction of meaningful data patterns from previous vaccine development programs, enabling companies to benefit from both successes and failures (Figure 3).
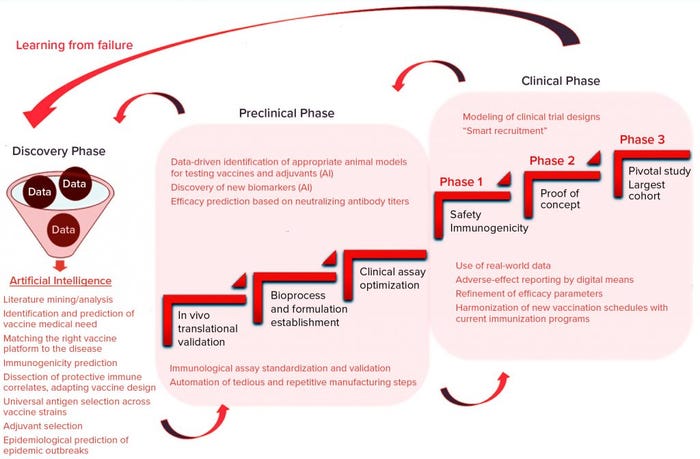
Figure 3: Application of AI within a vaccine development pipeline
Clinical Testing: The clinical stage is known to consume about half the budget invested into a drug company’s development pipeline (40). Retrospective analysis over the past 20 years found that roughly 66% of vaccines failed to succeed in licensure (41). In addition to the economic risks, vaccine clinical testing brings unique challenges. These products are tested in healthy individuals and often intended for a fragile pediatric population. Clinical evaluation of new vaccines can interfere with established vaccination programs and comorbidities, which could lead to unpredicted adverse reactions. Sporadic disappearances of infectious diseases even can make it difficult to test vaccine efficacy in a relevant population, further compromising the chances for success (42). Clearly, wide potential exists here to harness the strengths of AI and improve quality and efficiency of vaccine trials.
Conventional clinical trials follow a three-stage path (Figure 3), in which each phase gradually increases the number of participants to gain solid data that support an eventual MA. Iterative design based on computer modeling thus is a valuable alternative for modifying the course of a running trial dynamically in light of newly acquired data (43). Novel, flexible designs can help identify new predictive biomarkers of treatment efficacy through deep-system analysis of real-time data. Flexibility is key to advancing clinical developments for unmet medical needs (e.g., HIV vaccines), for which correlates of protective immune response remain elusive (44).
In silico modeling is extremely beneficial for fine tuning of data or dealing with data insufficiencies in clinical trials. Dorigatti et al. developed an algorithm to refine the efficacy of a Sanofi Pasteur vaccine against Dengue virus and discovered that the age of participants correlates with vaccine efficacy independently from their serostatus (45). Such applications of AI-powered ML analyses during trial execution could provide invaluable support in decision-making related to public and individual benefit-risk assessments (46).
Candidate selection and recruitment appear to be problematic too: About 86% of clinical trials fail to meet their recruitment timelines, which causes significant delays in execution. An automated search based on time-efficient mining of electronic health records could facilitate identification of the best trial candidates, efficiently replacing manual selection (40). AI also could help developers find the most favorable cohort by identifying homogeneous population subsets for responsiveness to a vaccine. Phase 1 enrollment can be complicated profoundly based on safety concerns relevant to healthy volunteers, an impact that should not be underestimated. That aspect is highly relevant for incurable diseases such as HIV acquired immune deficiency syndrome (AIDS). In that regard, digital options could be extremely helpful for educational purposes, helping to create a mature societal perception of participation in clinical trials (47).
To achieve an ultimate goal of eliminating a given infectious disease from society, vaccines usually are meant for use in large populations. And most prophylactic vaccines are intended for sensitive population cohorts (young children and the elderly). So it is not surprising that the safety bar will be much higher for vaccines than for drugs used, for example, in oncology.
Therefore, demonstration of vaccine safety and monitoring of adverse reactions are critical attributes of a vaccine portfolio. Thus, the US Food and Drug Administration (FDA) has created its Vaccine Adverse Event Reporting System (VAERS) platform to register, track, identify, and monitor all vaccine-associated adverse events (48). Note that natural language processing (NLP) tools already have been implemented for identification of adverse events related to Tdap vaccines for tetanus, diphtheria, and acellular pertussis vaccines (49). A pilot study has confirmed the feasibility of AI for adverse-event processing, which could reduce pharmacovigilance budgets significantly (50).
Implementing AI technologies into drug discovery will be beneficial only if they are accepted by a regulatory framework. Fortunately, the EU and US regulatory landscapes are being transformed gradually by the pressure of technology progress. In 2020, the European Commission published a white paper describing its strategy to catalyze AI integration into development of medicines (51). And of particular importance to clinical trials, another EU initiative funds the Data Analysis and Real-World Interrogation Network (DARWIN) project, which is intended to create a platform for access to real-world data (RWD) (52). That could facilitate patient enrollment into clinical trials and fill data gaps by pulling information from electronic health records and digital health devices.
In the United States, healthcare digital transformation began when the 21st Century Cures Act was signed into law in 2016 (53). Its intended benefits include expediting development of new advanced therapeutics and empowering clinical trials with RWD. In 2018, the FDA published guidance for sponsors on what types of information should be submitted to agencies when trials are designed based on computer simulations (54). To encourage the integration of AI tools into drug development, the agency’s Innovative Science and Technology Approaches for New Drugs (ISTAND) pilot program includes a strong focus on incorporating AI-based algorithms into patient diagnostics and clinical trial design (55).
Although those perspectives seem to be promising, no examples yet have been reported of successfully incorporating AI and RWD into clinical trial design (56). The lack of progress so far indicates that the industry is a long way from realizing that future state (56).
Infectious Disease Examples
In BPI’s next issue, we will conclude this review with a detailed discussion of how AI can be applied to specific vaccine targets: malaria, tuberculosis, HIV, herpesvirus, hepatitis, and pandemic coronavirus. Below, we highlight Zika virus as an preview of those discussions.
Zika: The biggest and most recent Zika virus outbreak happened in 2015 in South America. It was a major epidemic, with an estimated one million infected individuals. Zika is a mosquito-borne flavivirus that causes influenza-like clinical manifestations in most cases and neurological complications (Guillain–Barré syndrome) in a small proportion of adults. However, this latest epidemic has revealed an associated pathology in unborn children: congenital Zika syndrome (CZS), which occurs when women are infected during the first trimester of pregnancy. Recognition of such devastating fetal neurological damage led WHO to call for rapid vaccine development and label Zika as a global health threat (57).
Because the healthcare systems of countries where Zika is endemic are not prepared to respond to such epidemics, vaccine developers moved quickly to initiate and perform clinical trials. Before vaccine candidates could enter into a pivotal clinical trial stage, however, the epidemic had waned. Thus, no clinical trials evaluating Zika vaccine candidates have progressed beyond phase 2 (42). The unpredictable nature of Zika epidemics — together with a lack of successful pivotal efficacy studies — raises a global conundrum that leaves already overwhelmed healthcare systems in endemic countries unprepared for the next epidemic (58). In addition, some uncertainty remains about whether surrogate markers predicting protective immunity for other flavivirus vaccines can be extrapolated reliably to predict vaccine efficacy of candidate Zika vaccines (59).
We can expect AI technologies to be helpful in addressing several pitfalls of Zika vaccine development. One example of a short-time forecasting process for decision-making was documented in 2014 during an Ebola outbreak in Western Africa. After researchers determined that conventional clinical-trial designs for candidate Ebola vaccines would be unlikely to generate significant efficacy data, those trial designs were changed on an accelerated timeline (60–62). The most accurate long-term predictions are required to help strengthen awareness and preparedness of local healthcare systems far in advance of the next epidemic wave. Thus, strategies are most needed to apply AI’s potential in foreseeing upcoming epidemics in their infancies.
Unfortunately, knowledge gaps related to the poorly investigated Zika virus profoundly limit the possibility of fueling and training an AI model with relevant data. That problem is complicated further by the early developmental stage of available mathematical modeling approaches, which are not yet mature enough to generate real-world answers. Also at issue is the limited capacity of many public health agencies to adopt and implement such analytical approaches (63, 64).
Find more detailed discussion of how AI can help developers address some problems with Zika and other disease targets in part two of our review.
References
1 Lavecchia A. Deep Learning in Drug Discovery: Opportunities, Challenges and Future Prospects. Drug Disc. Today 24(10) 2019: 2017–2032; https://doi.org/10.1016/j.drudis.2019.07.006.
2 Yu K-H, Beam AL, Kohane IS. Artificial Intelligence in Healthcare. Nature Biomed. Eng. 2(10) 2018: 719–731; https://www.nature.com/articles/s41551-018-0305-z.
3 Xu M, David JM, Kim SH. The Fourth Industrial Revolution: Opportunities and Challenges. Int. J. Finan. Res. 9(2) 2018: 90–95; https://doi.org/10.5430/ijfr.v9n2p90.
4 Schwab K. The Fourth Industrial Revolution. Currency: New York, NY, 2017.
5 Marr B. The 9 Biggest Technology Trends That Will Transform Medicine and Healthcare in 2020. Forbes 1 November 2020; https://www.forbes.com/sites/bernardmarr/2019/11/01/the-9-biggest-technology-trends-that-will-transform-medicine-and-healthcare-in-2020/?sh=35032d5972cd.
6 Artificial Intelligence in Healthcare Market Global Forecast to 2026. Markets and Markets: Hadapsar, India, 2020.
7 Burki T. A New Paradigm for Drug Development. Lancet Dig. Health 2(5) 2020: e226; https://doi.org/10.1016/s2589-7500(20)30088-1.
8 Oyston P, Robinson K. The Current Challenges for Vaccine Development. J. Med. Microbiol. 61(7) 2012: 889–894; https://doi.org/10.1099/jmm.0.039180-0.
9 Immunization, Vaccines and Biologicals. World Health Organization: Geneva, Switzerland, 2020; https://www.who.int/teams/immunization-vaccines-and-biologicals.
10 Rappuoli R. Reverse Vaccinology. Curr. Opin. Microbiol. 3(5) 2000: 445–450; https://doi.org/10.1016/s1369-5274(00)00119-3.
11 Heaton PM. Challenges of Developing Novel Vaccines with Particular Global Health Importance. Front. Immunol. 11, 2020: 2456; https://doi.org/10.3389/fimmu.2020.517290.
12 Flower DR, et al. Computer Aided Selection of Candidate Vaccine Antigens. Immunome Res. 6(2) 2010: 1–16; https://dx.doi.org/10.1186%2F1745-7580-6-S2-S1.
13 He Y, et al. Emerging Vaccine Informatics. J. Biomed. Biotechnol. 2010: 218590; https://doi.org/10.1155/2010/218590.
14 Sunita, et al. Computational Tools for Modern Vaccine Development. Human Vacc. Immunother. 16(3) 2020: 723–735; https://doi.org/10.1080/21645515.2019.1670035.
15 Pennisi M, et al. Predicting the Artificial Immunity Induced By RUTI Vaccine Against Tuberculosis Using Universal Immune System Simulator (UISS). BMC Bioinformat. 20(6) 2019: 1–10; https://doi.org/10.1186/s12859-019-3045-5.
16 Doytchinova IA, Flower DR. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinformat. 8(1) 2007: 4; https://doi.org/10.1186/1471-2105-8-4.
17 Heinson AI, et al. Enhancing the Biological Relevance of Machine Learning Classifiers for Reverse Vaccinology. Int. J. Mol. Sci. 18(2) 2017: 312; https://doi.org/10.3390/ijms18020312.
18 Dalsass M, et al. Comparison of Open-Source Reverse Vaccinology Programs for Bacterial Vaccine Antigen Discovery. Front. Immunol. 10, 2019: 113; https://doi.org/10.3389/fimmu.2019.00113.
19 Ahmad TA, Eweida AE, Sheweita SA. B-Cell Epitope Mapping for the Design of Vaccines and Effective Diagnostics. Trials Vaccinol. 5, 2016: 71–83; https://doi.org/10.1016/j.trivac.2016.04.003.
20 Parvizpour S, et al. Epitope-Based Vaccine Design: A Comprehensive Overview of Bioinformatics Approaches. Drug Disc. Today 25(6) 2020: 1034–1042; https://doi.org/10.1016/j.drudis.2020.03.006.
21 Potocnakova L, Bhide M, Pulzova LB. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016: 6760830; https://doi.org/10.1155/2016/6760830.
22 Moise L, et al. The Two-Faced T Cell Epitope: Examining the Host-Microbe Interface with JanusMatrix. Human Vacc. Immunother. 9(7) 2013: 1577–1186; https://doi.org/10.4161/hv.24615.
23 Sharma M, et al. Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era. Vaccines 7(3) 2019: 89; https://doi.org/10.3390/vaccines7030089.
24 Russo G, et al. The Combination of Artificial Intelligence and Systems Biology for Intelligent Vaccine Design. Exp. Opin. Drug Disc. 15(11) 2020: 1267–1281; https://doi.org/10.1080/17460441.2020.1791076.
25 Bahrami AA, et al. Immunoinformatics: In Silico Approaches and Computational Design of a Multi-Epitope, Immunogenic Protein. Int. Rev. Immunol. 38(6) 2019: 307–322; https://doi.org/10.1080/08830185.2019.1657426.
26 Zikun Y, Bogdan P, Shahin N. An In Silico Deep Learning Approach to Multi-Epitope Vaccine Design: A SARS-CoV-2 Case Study. Scientif. Rep. 11(1) 2021: 3238; https://www.nature.com/articles/s41598-021-81749-9.
27 Chaudhury S, et al. Combining Immunoprofiling with Machine Learning to Assess the Effects of Adjuvant Formulation on Human Vaccine-Induced Immunity. Hum. Vacc. Immunother. 16(2) 2020: 400–411; https://doi.org/10.1080/21645515.2019.1654807.
28 Chaudhury S, et al. Identification of Immune Signatures of Novel Adjuvant Formulations Using Machine Learning. Scientif. Rep. 8(1) 2018: 1–11; https://www.nature.com/articles/s41598-018-35452-x.
29 Gonzalez-Dias P, et al. Methods for Predicting Vaccine Immunogenicity and Reactogenicity. Hum. Vacc. Immunother. 16(2) 2020: 269–276; https://dx.doi.org/10.1080%2F21645515.2019.1697110.
30 Golumbeanu M, et al. Combining Machine Learning and Mathematical Models of Disease Dynamics to Guide Development of Novel Disease Interventions. medRxiv 17 June 2021; https://doi.org/10.1101/2021.01.05.21249283.
31 Abbasi BA, et al. Identification of Vaccine Targets and Design of Vaccine Against SARS-CoV-2 Coronavirus Using Computational and Deep Learning Based Approaches. OSF Preprints 7 April 2020; https://osf.io/f8zyw.
32 Herati RS, Wherry EJ. What Is the Predictive Value of Animal Models for Vaccine Efficacy in Humans? Consideration of Strategies to Improve the Value of Animal Models. Perspect. Biol. 10(4) 2018: a031583; https://dx.doi.org/10.1101%2Fcshperspect.a031583.
33 Busquet F, et al. Harnessing the Power of Novel Animal-Free Test Methods for the Development of COVID-19 Drugs and Vaccines. Arch. Toxicol. 94(6) 2020: 2263–2272; https://dx.doi.org/10.1007%2Fs00204-020-02787-2.
34 Krewski D, et al. Toxicity Testing in the 21st Century: Progress in the Past Decade and Future Perspectives. Arch. Toxicol. 94(1) 2020: 1–58; https://doi.org/10.1007/s00204-019-02613-4.
35 Busquet F, et al. New European Union Statistics on Laboratory Animal Use: What Really Counts! ALTEX 37(2) 2020: 167–186; https://www.altex.org/index.php/altex/article/view/1755.
36 Luechtefeld T, et al. Machine Learning of Toxicological Big Data Enables Read-Across Structure Activity Relationships (RASAR) Outperforming Animal Test Reproducibility. Toxicol. Sci. 165(1) 2018: 198–212; https://doi.org/10.1093/toxsci/kfy152.
37 Zhang Q, et al. Molecular Signaling Network Motifs Provide a Mechanistic Basis for Cellular Threshold Responses. Environ. Health Perspect. 122(12) 2014: 1261–1270; https://dx.doi.org/10.1289%2Fehp.1408244.
38 Li Z, et al. Dose-Response Modeling of Etoposide-Induced DNA Damage Response. Toxicol. Sci. 137(2) 2014: 371–384; https://doi.org/10.1093/toxsci/kft259.
39 Ng’uni T, Chasara C, Ndhlovu ZM. Major Scientific Hurdles in HIV Vaccine Development: Historical Perspective and Future Directions. Front. Immunol. 11, 2020: 2761; https://doi.org/10.3389/fimmu.2020.590780.
40 Harrer S, et al. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci. 40(8) 2019: 577–591; https://doi.org/10.1016/j.tips.2019.05.005.
41 Lo AW, Siah KW, Wong CH. Report 0898-2937: Estimating Probabilities of Success of Vaccine and Other Anti-Infective Therapeutic Development Programs. National Bureau of Economic Research: Cambridge, MA, 2020; https://doi.org/10.1162/99608f92.e0c150e8.
42 Pattnaik A, Sahoo BR, Pattnaik AK. Current Status of Zika Virus Vaccines: Successes and Challenges. Vaccines 8(2) 2020: 266; https://dx.doi.org/10.3390%2Fvaccines8020266.
43 Angus DC, et al. Adaptive Platform Trials: Definition, Design, Conduct, and Reporting Considerations. Nature Rev. Drug Discov. 18(10) 2019: 797–808; https://doi.org/10.1038/s41573-019-0034-3.
44 Richert L, et al. Accelerating Clinical Development of HIV Vaccine Strategies: Methodological Challenges and Considerations in Constructing an Optimised Multi-Arm Phase I/II Trial Design. Trials 15(1) 2014: 1–12; https://doi.org/10.1186/1745-6215-15-68.
45 Dorigatti I, et al. Refined Efficacy Estimates of the Sanofi Pasteur Dengue Vaccine CYD-TDV Using Machine Learning. Nature Comm. 9(1) 2018: 1–9; https://www.nature.com/articles/s41467-018-06006-6.
46 Black S, et al. Transforming Vaccine Development. Semin. Immunol. 50, August 2020: 101413; https://doi.org/10.1016/j.smim.2020.101413.
47 Buckingham L, et al. Going Social: Success in Online Recruitment of Men Who Have Sex with Men for Prevention HIV Vaccine Research. Vaccine 35(27) 2017: 3498–3505; https://dx.doi.org/10.1016%2Fj.vaccine.2017.05.002.
48 Zhou W, Ellenberg SS. Surveillance for Safety After Immunization: Vaccine Adverse Event Reporting System (VAERS) — Morbidity and Mortality Weekly Report. MMWR Surveillance Summaries Surveillance Summaries. US Centers for Disease Control: Atlanta, GA, 2003; https://www.cdc.gov/mmwr/preview/mmwrhtml/ss5201a1.htm.
49 Zheng C, et al. The Use of Natural Language Processing to Identify Tdap-Related Local Reactions at Five Health Care Systems in the Vaccine Safety Datalink. Int. J. Med. Informat. 127, 2019: 27–34; https://doi.org/10.1016/j.ijmedinf.2019.04.009.
50 Schmider J, et al. Innovation in Pharmacovigilance: Use of Artificial Intelligence in Adverse Event Case Processing. Clin. Pharmacol. Therapeut. 105(4) 2019: 954–961; https://doi.org/10.1002/cpt.1255.
51 EMA Regulatory Science to 2025. European Medicines Agency: Amsterdam, The Netherlands, 2020; https://www.ema.europa.eu/en/documents/regulatory-procedural-guideline/ema-regulatory-science-2025-strategic-reflection_en.pdf.
52 EU Big Data Stakeholder Forum: Summary Report from the Big Data Stakeholder Forum Held on 15/12/2020. European Medicines Agency: Amsterdam, The Netherlands, 2020; https://www.ema.europa.eu/en/documents/report/summary-report-eu-big-data-stakeholder-virtual-forum_en.pdf.
53 21st Century Cures Act. US Public Law 114–255, 13 December 2016; https://www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act.
54 CBER, CDER. Adaptive Design Clinical Trials for Drugs and Biologics: Guidance for Industry. US Food and Drug Administration: Rockville, MD, 2019; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/adaptive-design-clinical-trials-drugs-and-biologics-guidance-industry.
55 Innovative Science and Technology Approaches for New Drugs (ISTAND) Pilot Program. US Food and Drug Administration: Rockville, MD, 2021; https://www.fda.gov/drugs/drug-development-tool-ddt-qualification-programs/innovative-science-and-technology-approaches-new-drugs-istand-pilot-program.
56 Shah P, et al. Artificial Intelligence and Machine Learning in Clinical Development: A Translational Perspective. NPJ. Dig. Med. 2(1) 2019: 1–5; https://doi.org/10.1038/s41746-019-0148-3.
57 WHO/UNICEF. Zika Virus (ZIKV) Vaccine Target Product Profile (TPP):Vaccine to Protect Against Congenital Zika Syndrome for Use During an Emergency. World Health Organization: Geneva, Switzerland, February 2017; https://www.who.int/publications/m/item/who-unicef-zika-virus-(zikv)-vaccine-target-product-profile-(tpp).
58 Vannice KS, et al. Demonstrating Vaccine Effectiveness During a Waning Epidemic: A WHO/NIH Meeting Report on Approaches to Development and Licensure of Zika Vaccine Candidates. Vaccine 37(6) 2019: 863–868; https://doi.org/10.1016/j.vaccine.2018.12.040.
59 Barrett AD. Current Status of Zika Vaccine Development: Zika Vaccines Advance into Clinical Evaluation. npj Vaccines 3(1) 2018: 1–4; https://www.nature.com/articles/s41541-018-0061-9.
60 George DB, et al. Technology to Advance Infectious Disease Forecasting for Outbreak Management. Nature Comm. 10(1) 2019: 1–4; https://www.nature.com/articles/s41467-019-11901-7.
61 Zhao S, et al. Simple Framework for Real-Time Forecast in a Data-Limited Situation: The Zika Virus (ZIKV) Outbreaks in Brazil from 2015 to 2016 As an Example. Paras. Vect. 12(1) 2019: 1–13; https://doi.org/10.1186/s13071-019-3602-9.
62 Akhtar M, Kraemer MU, Gardner LM. A Dynamic Neural Network Model for Predicting Risk of Zika in Real Time. BMC Med. 17(1) 2019: 1–16; https://doi.org/10.1186/s12916-019-1389-3.
63 Chowell G, et al. Using Phenomenological Models to Characterize Transmissibility and Forecast Patterns and Final Burden of Zika Epidemics. PLoS Curr. 8, 31 May 2016; https://doi.org/10.1371/currents.outbreaks.f14b2217c902f453d9320a43a35b9583.
64 Kobres P-Y, et al. A Systematic Review and Evaluation of Zika Virus Forecasting and Prediction Research During a Public Health Emergency of International Concern. PLoS Neglect. Trop. Dis. 13(10) 2019: e0007451; https://doi.org/10.1371/journal.pntd.0007451.
Aleksandra Nevmerzhitskaya is a consultant, Mariya Gromova is a senior consultant, and Dr. Michael Pfleiderer is a principal consultant at Biopharma Excellence (a PharmaLex company), Munich Technology Center, Agnes-Pockels-Bogen 1, 80992 Munich, Germany; [email protected].
You May Also Like





