Harnessing the Untapped Power of the Human Commensal ViromeHarnessing the Untapped Power of the Human Commensal Virome
October 20, 2022
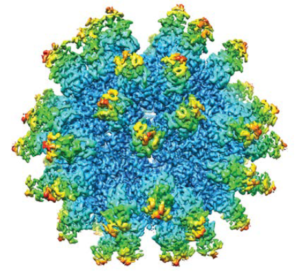
Anelloviruses are icosahedral virions that encapsdate small single-stranded DNA genomes. (HTTPS://RINGTX.COM)
Viruses are ubiquitous members of every ecosystem, including the human body (1). A common misconception is that all viruses are harmful. In fact, their relationships to human hosts can be pathogenetic, symbiotic, or commensal. In the latter case, a virus benefits from but neither harms nor helps its host (2). Commensal viruses are as abundant as human cells, and their coexistence has persisted for millennia, permitting our immune systems to recognize them as part of us. Among the most abundant members of the human commensal virome are anelloviruses (3). The Anelloviridae family is believed to exist inside every healthy human being from birth, having been shared maternally through placental blood, and such viruses persist throughout a human host’s life (4–6).
Anelloviridae comprises more than 30 genera (7) characterized by icosahedral virions that encapsidate a small single-stranded DNA (ssDNA) genome (8). Anello is Latin for “ring.” Anelloviruses are so named for their circular genomes. Most healthy humans are infected with unique lineages of anelloviruses that make up an individual’s “anellome,” and such viruses have been present in nearly every human tissue that researchers have investigated (9–12). This family of commensal viruses remained highly understudied for decades because of their lack of association to human disease (5) and their inability to culture — until a small group of researchers began investigating biological characteristics that could make anelloviruses highly valuable to medicine.
The Unique Biology of Anelloviruses
Anelloviruses were first discovered in humans in 1997 (13). Since then, academic researchers uncovered the family’s expanded diversity and host range (6, 14–18). No evidence of a relationship to human disease has been observed. Anelloviruses proved difficult to culture, limiting early research to genomic sequencing on their diversity, distribution, and tropism. Extensive sequencing in healthy individuals over time has shaped a new perspective of anelloviruses as integral components of a healthy human virome (6, 18, 19).
Diversity: Targeted sequencing has revealed that anelloviruses have few constraints on their evolution and transmission, allowing for significant diversity within an individual’s anellome (6). Anelloviral diversity even is three to four times broader than that of other human viruses, including adenoassociated viruses (AAVs). Anelloviruses have been detected across the animal kingdom, but to date only three genera have been observed to infect humans chronically: alphatorquevirus, betatorquevirus, and gammatorquevirus. Among those, alphatorqueviruses are the most abundant lineage. The extensive diversity of anelloviruses is unique to each individual and highly stable over time (6, 19).
Persistence: Early evidence of anellovirus transmission through blood transfusion (20) sparked a large-scale investigation into the kinetics and transmissibility of anelloviruses within and between individuals (6). Results from such studies indicate that an individual’s anellome can be transmitted to and persist for many months in other humans, seamlessly assimilating with a recipient’s own anellome (6). Subsequent research has detected specific anellovirus lineages in subject blood samples for over 30 years (19), suggesting long-term stability of anelloviruses in humans.
Tropism: Anelloviruses have been identified in heart, muscle, retina, and many other tissue types. This naturally broad tropism was uncovered through extensive genomic sequencing across the human body (9–12). A 2022 study showed that anellovirus isolated from retinal tissue and injected into a mouse exhibited tropism to the tissue from which it was isolated (21). Those results suggest that anellovirus capsids have properties that enable tissue-specific tropism.
Nonintegration with Host Genome: A critical characteristic of anelloviruses is that their genomes remain as episomes within host-cell nuclei (6). This nonintegration eliminates risks for insertional mutagenesis, and in nondividing cells, episomes can remain alongside the nucleus for the life of the cell.
Immune Stealth: Anelloviruses are associated with no human disease and have lived alongside our immune system for millennia, suggesting a lack of immune system engagement with anellovirus infection. In the blood transfusion studies previously mentioned, recipients have shown no pathogenic responses to donor anellomes despite being “co-infected” by them (6). Using a comprehensive tool for anellovirus antibody analysis, researchers have shown that natural anelloviruses are even highly immune favorable compared with other human viruses (22).
This immune stealth remained enigmatic until the discovery of unique viral structures. High-resolution images of anelloviral capsids taken by researchers at my company, Ring Therapeutics, have revealed 60 open reading frame 1 (ORF1) protein fragments that feature a modified jelly-roll–domain core architecture, with residues that extend from the surface to form a novel elongated surface structure in which hypervariable regions lie at the apex (23). Five of those surface proteins pack together in a ringed structure resembling a crown. At the apex of the anellovirus crown is a domain formed by the hypervariable region that is hypothesized to hinder antibody binding sterically to key ORF1 motifs. Such steric effects could prevent antibody recognition, enabling anelloviruses to infect humans repeatedly while evading the immune system. Further research is needed to investigate this hypothesis.
We have leveraged the unique properties of natural anelloviruses to engineer a class of virally vectored treatments that we call AnelloVector therapeutics. Our synthetic vectors are designed to exhibit the same characteristics as wild-type anelloviruses (6, 21–23). Therapeutics delivered by these vectors have potential to be safe, redosable, and potent, helping to usher in new approaches to gene therapy and programmable medicine more generally.
Making AnelloVector Therapeutics
AnelloVector therapeutics were designed using a wealth of new information on anelloviruses that could help to transform commensal viruses into effective therapies. Prior difficulties with culture processes had discouraged work with anelloviruses. However, a 2022 study has showcased a successful in vitro system for anellovirus production, opening the door for deeper investigations (21).
From demonstrating the shear diversity and abundance of anelloviruses in the human body to increasing our understanding of anellovirus structure, Ring seeks to advance anellovirus research to harness the virus’s properties for therapeutic applications. The company’s capture-sequencing technology has helped to construct the world’s largest collection of genomic data about human-derived anelloviruses (6). In 2021, the database contained more than 5,000 viral sequences, and that number continues to grow (6). Mining those data has begun to reveal key insights, most notably that the ORF1 capsid protein defines critical aspects of anellovirus biology.
Researchers have confirmed experimentally that the ORF1 gene encodes the corresponding capsid protein (21, 23). They also have screened several anellovirus peptides (e.g., those associated with ORF1, ORF2, ORF3, and torque teno virus apoptosis-inducing protein (TAIP)) against human sera to detect potential immune reactivity. The vast majority (~85%) of screened materials showed no reaction, and the few antibody-reactive peptides were confined largely to the C-terminal region of ORF1 (22). However, when ORF1 was expressed in cell lines to produce a viral particle, ORF1 exhibited proteolysis of the C-terminal region, showing that the immunogenic region of ORF1 may not be essential for particle formation (23).
Once genomic predictions are validated through screening of anellovirus characteristics in vitro, those data can be fed back into a machine learning system to design generations of anelloviruses for enhanced vector characteristics such as tissue-specific tropism, redosability, and potency without immunogenicity. One such anellovirus whose genome was isolated from human retinal epithelium was synthesized and cultured (21). Then the recombinant virus was reintroduced to its tissue of origin, where it demonstrated even higher and more efficient specific infectivity than what can be achieved using AAVs, providing proof of principle for application of AnelloVector therapeutics (21).
Now that the building blocks of anelloviruses are understood better than ever before, there is significant potential to design anelloviruses for delivery of nonnative payloads for a breadth of indications. Payload versatility coupled with potential for scalable manufacturing could make anellovirus a promising platform for programmable medicine based on the commensal virome.
References
1 Harris HMB, Hill C. A Place for Viruses on the Tree of Life. Front. Microbiol. 11, 2021: 604048; https://doi.org/10.3389/fmicb.2020.604048.
2 Liang G, Bushman F. The Human Virome: Assembly, Composition, and Host Interactions. Nature Rev. Microbiol. 19(8) 2021: 514–527; https://doi.org/10.1038/s41579-021-00536-5.
3 Virgin HW, Wherry EJ, Ahmed R. Redefining Chronic Viral Infection. Cell 138(1) 2009: 30–50; https://doi.org/10.1016/j.cell.2009.06.036.
4 Tyschik EA, et al. Torque Teno Virus Dynamics During the First Year of Life. Virol. J. 15(1) 2018: 96; https://doi.org/10.1186/s12985-018-1007-6.
5 Koonin EV, Dolja VV, Krupovic M. The Healthy Human Virome: From Virus–Host Symbiosis to Disease. Curr. Opin. Virol. 47, 2021: 86–94; http://dx.doi.org/10.1016/j.coviro.2021.02.002.
6 Arze CA, et al. Global Genome Analysis Reveals a Vast and Dynamic Anellovirus Landscape Within the Human Virome. Cell Host & Microbe 29(8) 2021: 1305–1315; https://doi.org/10.1016/j.chom.2021.07.001.
7 Virus Taxonomy: 2020 Release. International Committee on Taxonomy of Viruses, March 2021; https://ictv.global/taxonomy/history.
8 Takahashi K, et al. Very High Prevalence of TT Virus (TTV) Infection in General Population of Japan Revealed by a New Set of PCR Primers. Hepatol. Res. 12(3) 1998: 233–239; https://doi.org/10.1016/S1386-6346(98)00068-0.
9 Kaczorowska J, van der Hoek L. Human Anelloviruses: Diverse, Omnipresent, and Commensal Members of the Virome. FEMS Microbiol. Rev. 44(3) 2020: 305–313; https://doi.org/10.1093/femsre/fuaa007.
10 Hijikata M, Takahashi K, Mishiro S. Complete Circular DNA Genome of a TT Virus Variant (Isolate Name SANBAN) and 44 Partial ORF2 Sequences Implicating a Great Degree of Diversity Beyond Genotypes. Virology 260(1) 1999: 17–22; https://doi.org/10.1006/viro.1999.9797.
11 Okamoto H, et al. Species-Specific TT Viruses in Humans and Nonhuman Primates and Their Phylogenetic Relatedness. Virology 277(2) 2020: 368–378; https://doi.org/10.1006/viro.2000.0588.
12 Takahashi K, et al. Identification of a New Human DNA Virus (TTV-Like Mini Virus, TLMV) Intermediately Related to TT Virus and Chicken Anemia Virus. Arch. Virol. 145(5) 2000: 979–993; https://doi.org/10.1007/s007050050689.
13 Nishizawa T, et al. A Novel DNA Virus (TTV) Associated with Elevated Transaminase Levels in Posttransfusion Hepatitis of Unknown Etiology. Biochem. Biophys. Res. Comm. 241(1) 1997: 92–97; https://doi.org/10.1006/bbrc.1997.7765.
14 Biagini P, et al. Circular Genomes Related to Anelloviruses Identified in Human and Animal Samples by Using a Combined Rolling-Circle Amplification/Sequence-Independent Single Primer Amplification Approach. J. General Virol. 88(10) 2007: 2696–2701; https://doi.org/10.1099/vir.0.83071-0.
15 Spandole S, et al. Human Anelloviruses: An Update of Molecular, Epidemiological, and Clinical Aspects. Arch. Virol. 160(4) 2015: 893–908; https://doi.org/10.1007/s00705-015-2363-9.
16 de Souza WM, et al. Discovery of Novel Anelloviruses in Small Mammals Expands the Host Range and Diversity of the Anelloviridae. Virology 514, 2018: 9–17; http://doi.org/10.1016/j.virol.2017.11.001.
17 Varsani A, et al. Taxonomic Update for Mammalian Anelloviruses (Family Anelloviridae). Arch. Virol. 166(10) 2021: 2943–2953; https://doi.org/10.1007/s00705-021-05192-x.
18 Cebriá-Mendoza M, et al. Deep Viral Blood Metagenomics Reveals Extensive Anellovirus Diversity in Healthy Humans. Sci. Rep. 11, 2021: 6921; https://doi.org/10.1038/s41598-021-86427-4.
19 Kaczorowska J, et al. Diversity and Long-Term Dynamics of Human Blood Anelloviruses. J. Virol. 96(11) 2022: https://doi.org/10.1128/jvi.00109-22.
20 Kapoor A, et al. Virome Analysis of Transfusion Recipients Reveals a Novel Human Virus That Shares Genomic Features with Hepaciviruses and Pegiviruses. mBio 6(5) 2015: e01466-15; https://doi.org/10.1128/mBio.01466-15.
21 Nawandar DM, et al. Human Anelloviruses Produced by Recombinant Expression of Synthetic Genomes. bioRxiv 28 April 2022; https://doi.org/10.1101/2022.04.28.489885.
22 Venkataraman T, et al. Comprehensive Profiling of Antibody Responses to the Human Anellome Using Programmable Phage Display. bioRxiv 29 March 2022; https://doi.org/10.1101/2022.03.28.486145.
23 Liou S-h, et al. Anellovirus Structure Reveals a Mechanism for Immune Evasion. bioRxiv 2 July 2022; https://doi.org/10.1101/2022.07.01.498313.
Tuyen Ong, MD, MBA, is chief executive officer partner at Flagship Pioneering and chief executive officer of Ring Therapeutics, 620 Memorial Drive, Cambridge, MA 02139; https://ringtx.com/contact.
You May Also Like





