Adenovirus Downstream Process Intensification: Implementation of a Membrane AdsorberAdenovirus Downstream Process Intensification: Implementation of a Membrane Adsorber
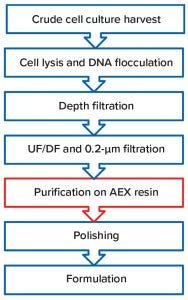
Figure 1: Adenovirus downstream process
Historically, companies developing vaccines have used attenuated pathogens, inactivated infectious agents, or antigenic constituents purified from pathogenic sources. In the past 20 years, technological advances such as recombination and viral vectors, have enabled development of vaccines against diseases with previously no available treatments (1).
Viral vectors have become one of the most rapidly evolving and promising fields in vaccinology and regenerative medicine. In addition to preventing infectious disease, they have a broad range of potential applications, including treatment of hereditary genetic disorders and cancers.
Adenoviruses are the most common vectors used to develop recombinant vaccines. They allow for development of vaccines against highly infectious and pathogenic viruses that would be too dangerous or technically difficult to produce at an industrial scale (2).
Vaccine developers modify the adenoviral vectors to express antigens from specific pathogens once the vectors enter target cells of a vaccinated individual. Those antigens then elicit a specific immune response against the desired pathogen.
Adenoviruses can be manufactured as shown in Figure 1. First, a human embryonic kidney (HEK293) cell culture is infected to produce the recombinant adenovirus. Cells are lysed, and their DNA is flocculated before clarification by depth filtration. An ultrafiltration/diafiltration step using a large cut-off membrane then removes contaminants such as soluble host-cell proteins and small DNA fragments. A resin-based anion-exchange (AEX) chromatography unit operation is performed in capture mode before a polishing step reduces residual DNA to trace levels before final formulation of the viral-vector product.

Photo 1: Comparing facility footprints of membrane and resin processes
With growing competition in the vaccine industry, manufacturers need to maximize their process productivity and optimize costs while ensuring strong product quality. To achieve that end, we considered replacing the resin-based AEX (Figure 1, in red) with a single-use membrane adsorber. Membrane technology offers numerous advantages over resins in virus-capture steps (3–5). The large pore size of membrane adsorbers provides maximum ligand accessibility for large particles and eliminates the need for diffusion through resin pores. Consequently, membranes allow for flow rates that are typically 10-fold higher than with classical resins and boost viral-capture capacity 5–20 times greater than that of column-based processes. Because no column packing or unpacking is required, membrane adsorbers are easy to use. They have a small footprint and are fully disposable, which reduces the risk of cross contamination and lowers both validation costs and environmental impact (6).
For successful process intensification, we wanted a membrane adsorber that would perform better than the resin. The challenge for the membranes we tested was a well-established AEX resin capture step in our existing process that gave high virus recovery and high reduction of host-cell contaminants. Thus, the benchmark for this step was a capacity of >1.5 × 1012 viral particles (VP)/mL, a minimum yield of 90%, more than three logs reduction in host-cell DNA, and two logs reduction in host-cell proteins (HCPs).
We assessed three Q-type membrane adsorbers from three different manufacturers in parallel at small scale under comparable conditions for load, capacity, and elution. Eventually we chose the Sartobind Q membrane for implementation in our downstream process, then evaluated its performance for process intensification.
Materials and Methods
Laboratory-Scale Experiments: Small-scale experiments used a 20-mL Q resin (100-mm packed bed height) and a Sartobind Q Nano (3-mL membrane, 8-mm bed height) from Sartorius Stedim Biotech. Adenoviral load material was identical for all small-scale experiments conducted (aliquots stored at –70 °C).
For the first experiment, we compared the resin and membrane adsorber directly using an existing protocol developed for the resin. This involved sanitizing the column and membrane adsorber with sodium hydroxide (NaOH), equilibrating with saline Tris buffer, and loading with 1.7 × 1012 VP/mL of static phase. We used a flow rate of 10 mL/min in both cases (3.33 MV/min for membrane and 0.5 CV/min for resin). A wash with equilibration buffer was followed by elution with Tris buffer and increased NaCl concentration. Both resin and membrane were regenerated with NaOH and stored under conditions recommended by their suppliers.
We performed several optimization runs to test different pH levels and different NaCl concentrations for loading, wash, and elution. Finally, the maximal dynamic binding capacity (DBC) was determined for different loading buffers containing low and high salt concentrations.
Implementation in a Pilot-Scale Process: Our scale-up trial used a Sartobind Q capsule of 150 mL (8-mm bed height) loaded with a freshly prepared adenovirus suspension. First we sanitized the membrane with NaOH, then flushed it with saline Tris solution (NaCl 2 M) before equilibration. We loaded 1.5 × 1013 virus particles in suspension per milliliter of membrane at 3.33 MV/min (0.5 L/min). No regeneration step was performed, and the membrane was discarded after use. Then we completed the downstream process to produce our drug substance.
Analytical Assays: We quantified adenovirus particles in the different fractions using an AEX high-performance liquid chromatography (HPLC) assay. We based our virus particle recovery percentage calculation on the total amount of virus in the load fraction compared with that recovered in the eluate. HCPs were measured with an in-house enzyme-linked immunosorbent assay (ELISA). Host-cell DNA was quantified with two quantitative polymerase chain reaction (QPCR) assays: one for the total amount of DNA and one specifically for larger DNA molecules.
Results
Laboratory-Scale Comparison of the AEX Resin with a Membrane Adsorber: Our first set of experiments directly compared the resin and membrane using the same protocol developed for the resin. Figure 2a shows UV profiles (280 nm) of chromatograms for the resin, and Figure 2b shows those for the membrane. In both cases, we observed flowthrough of unbound proteins but no breakthrough of virus. Step elution using a buffer with a higher NaCl concentration resulted in elution of bound viruses, and the last peak resulted from stripping with 2M NaCl. These results suggested that the membrane adsorber functioned similarly to the AEX resin.
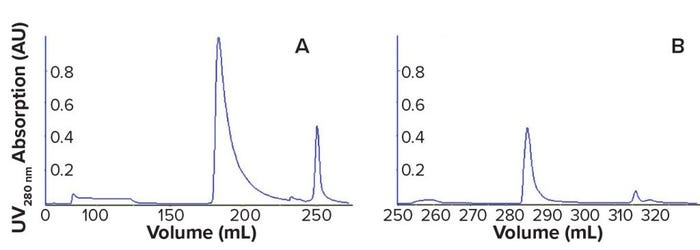
Figure 2: UV 280-nm chromatograms from reference resin (a) and Sartobind membrane (b); experiment was conducted with a 20-mL Q resin column and a 3-mL Sartobind Q (8-mm bed height). Resin and membrane first were sanitized with NaOH 1 M, then equilibrated with Tris-NaCl buffer. Virus load was 1.7 × 1012 virus particles (VP)/mL, and step elution used the same Tris buffer with increased NaCl concentration.
We concluded that conditions used for the resin could be applied directly to the membrane adsorber. Our next step was to determine whether the membrane could outperform the resin by showing higher capacity at comparable impurity removal.
Impact of Loading Conditions on Membrane Performance at Laboratory Scale: In the next set of experiments, we evaluated the effects of different pH levels and NaCl concentrations on virus binding and overall purity at constant load. We found pH to have no major impact (data not shown), so we minimally changed the original protocol for that factor.
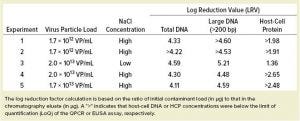
Table 1: Comparing host-cell DNA and protein reductions by resin (experiment 1) and membrane adsorber in different load conditions (experiments 2–5)
Next we determined DBC of the membrane adsorber under two different loading conditions with low and high salt concentrations. Those conditions are known to enable virus binding (6).
We overloaded the membrane with adenovirus at high NaCl concentration until breakthrough was observed. With a low NaCl concentration, all available material was loaded without reaching virus breakthrough. We determined DBCs to be 12–25× higher for the membrane than for the resin depending on the NaCl concentration: The low salt concentration gave a DBC of >3.75 × 1013 VP/mL compared to ~2 × 1013 VP/mL at high salt concentration. However, direct comparison of loading at high and low salt concentrations revealed a negative impact on HCP removal in the latter (Table 1, experiment 3). In fact, low salt concentration in the load sample favored binding of HCP to the membrane adsorber and coelution with the viral-vector product. By contrast, the higher salt concentration prevented binding of HCP to the membrane, thus preventing contamination of the product during elution. Thus, HCP reduction at high salt conditions was comparable between the Sartobind Q membrane and the reference resin, even when the membrane was loaded with 10- to 12-fold more adenovirus (Table 1).
DNA clearance was the same for both the membrane adsorber and the resin reference under the same operating conditions. The membrane’s ability to remove >4 log of DNA was confirmed in all conditions, even when throughput (the volume of virus suspension loaded per milliliter of membrane) was increased >10-fold (Table 1).
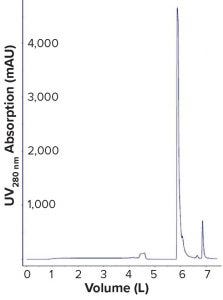
Figure 3: Chromatogram of Sartobind membrane at pilot scale; experiment was conducted with a 150-mL Sartobind Q (8-mm bed height) at 3.33 MV/min. The membrane first was sanitized with NaOH 1 M, then equilibrated with saline Tris buffer. Virus load was 1.7 × 1013 VP/mL of membrane, and elution used the same Tris buffer and a higher NaCl concentration.
Scaling Up Adenovirus Membrane Chromatography Purification: Based on the results from our laboratory-scale experiments, we decided to scale up the membrane adsorber by implementing a 150-mL membrane in a pilot-scale process. Scale-up was straightforward: We kept bed height constant and maintained the same flow rate per volume of membrane as at small scale. Adenovirus was loaded at the capacity of 1.7 × 1013 VP/mL of membrane. We processed the virus suspension from tangential-flow filtration (TFF) on the Sartobind Q 150-mL at a flow rate of 3.33 MV/min (1.66 MV/mL for sanitization). Photo 1 shows a footprint comparison between the membrane adsorber and the resin at this scale. No stainless-steel holder was required for the membrane. Figure 3 shows the chromatogram obtained during our pilot-scale run.
The chromatography profile and results obtained with the 150-mL membrane were consistent with data obtained from laboratory-scale experiments. Table 2 lists results of host-cell DNA and HCP removal for the membrane step at pilot scale. Product quality in the membrane eluate met target specifications, with HCPs below the quantification level, DNA reduced by a log reduction value (LRV) of >4, and product recovery ~90%. Moreover, the overall process yield (in number of doses) and quality (based on HCP and host-cell DNA levels) of the drug substance produced with the membrane adsorber were similar to those of the original process using an AEX column at pilot scale. These results show that the membrane adsorber performed as required at pilot scale.

Table 2: Host-cell protein and DNA reduction with Sartobind Q 150-mL in a pilot-scale process
Projection to 200-L Manufacturing Scale: To assess the impact and potential benefits of replacing the chromatography column at large manufacturing scale with a membrane adsorber, we calculated projections based on the pilot-scale experience described above. Table 3 list the results of this projection to 200-L scale. An 8-L column packed with resin can be replaced by a 0.8-L Sartobind Q membrane adsorber, reducing facility footprint, buffer consumption, and processing time. Furthermore, the preparation time for the step is reduced by elimination of the column-packing step. Alternatively, replacing the AEX resin with a 0.15-L membrane adsorber and cycling that device six times could allow the step to be performed in an even smaller footprint and at even lower cost but with greater productivity than with a column. Thus, we believe that membrane adsorbers are an effective tool to enhance the productivity of biopharmaceutical processes.
An Intensified Process
We have studied the possibility of replacing a Q resin chromatography column in our current adenovirus downstream process with a membrane adsorber. The results provide evidence that the Sartobind Q membrane adsorber in a bind-and-elute purification step would be suitable as part of our platform process for adenovirus manufacturing. Relatively little development work was required to enable this change.
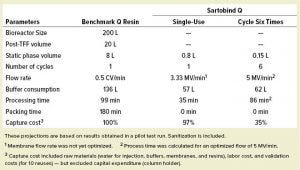
Scale-up projections to 200-L manufacturing scale; TFF = tangential-flow filtration
Replacing the resin with a membrane adsorber could increase capacity of the chromatography step by over 10-fold without compromising impurity removal. The increase in capacity entails a number of beneficial effects: The smaller footprint makes equipment handling more user friendly, which is especially beneficial for large-scale operations. The smaller membrane allowed for 58% less buffer consumption (hence, less waste) compared with the column process. Moreover, with the ready-to-use membrane adsorber, column packing and validation are eliminated, as are cleaning and related validation of the device. All those benefits can lead to significant time and cost savings compared with traditional AEX resin chromatography.
Data shown herein indicate that implementing membrane adsorber technology in the adenovirus downstream process could provide the targeted process intensification. Residual levels of HCP and host-cell DNA fall within the specifications for the drug substance and enable further implementation in clinical manufacturing.
Acknowledgments
We thank Fabian Delfosse (employed by GSK at the time of the study) for laboratory assistance. This work was sponsored by GlaxoSmithKline Biologicals SA, which was involved in all stages of study conduct and analysis. Authors Abrecht and Permanne are employees of the GSK group of companies; Permanne reports ownership of shares and/or restricted shares in GSK. Authors Pressac and Boulais are employees of Sartorius Stedim Biotech, manufacturer of the Sartobind Q membrane.
References
1 Nascimento IP, Leite LCC. Recombinant Vaccines and the Development of New Vaccine Strategies. Braz. J. Med. Biolog. Res. 45(12) 2012: 1102–1111; doi:10.1590/S0100-879X2012007500142.
2 Wold WS, Toth K. Adenovirus Vectors for Gene Therapy, Vaccination, and Cancer Gene Therapy. Curr. Gene Ther. 13(6) 2013: 421–433.
3 Vicente T, et al. Anion-Exchange Membrane Chromatography for Purification of Rotavirus-Like Particles. J. Membrane Sci. 311(1) 2008: 270–283; doi:10.1016/j.memsci.2007.12.021.
4 Vicente T, et al. Large-Scale Production and Purification of VLP-Based Vaccines. J. Invert. Pathol. 107, 2011: S42–S48; doi:10.1016/j.jip.2011.05.004.
5 Weigel T, et al. A Membrane-Based Purification Process for Cell Culture-Derived Influenza A Virus. J. Biotechnol. 220, 20 February 2016: 12–20; doi:10.1016/j.jbiotec.2015.12.022.
6 Boulais A, et al. Enabling Viral Vaccine Production: Implementing Process-Scale Adenovirus Purification with a Single-Use Platform. Gen. Eng. Biotechnol. News 36(20) 2016.
7 Madabhushi SR, et al. Quantitative Assessment of Environmental Impact of Biologics Manufacturing Using Process Mass Intensity Analysis. Biotechnol. Prog. 34(6) 2018: 1566–1573; doi:10.1002/btpr.2702.
Helge Abrecht is an expert scientist, and Philippe Permanne is a senior scientist, both in cell and viral drug substance analytical development at GlaxoSmithKline in Rixensart, Belgium. Geoffrey Pressac is an application specialist in separation technologies, and Amélie Boulais is vaccine platform marketing manager, both at Sartorius Stedim France S.A.S., Aubagne, France. Abrecht, Pressac, and Boulais conceived and designed the study reported herein; Abrecht and Pressac acquired the data, analyzed them, and interpreted the results; Abrecht, Pressac, Boulais, and Permanne drafted or critically revised the manuscript for important intellectual content. All authors had full access to the data and approved the manuscript before it was submitted by the corresponding author.
You May Also Like





