Analysis of Trace-Level, High-Risk HCPs: Proteomics Advances for Preventing Degradation of Polysorbates in Biotherapeutic FormulationsAnalysis of Trace-Level, High-Risk HCPs: Proteomics Advances for Preventing Degradation of Polysorbates in Biotherapeutic Formulations
milliporesigma (https://www.emdmillipore.com)
Polysorbate-80 (PS-80) and polysorbate-20 (PS-20) are used widely in formulation of biotherapeutic products for preventing surface adsorption and as stabilizers against protein aggregation (1). Degradation of polysorbates can cause turbidity and potential formation of subvisible particles mainly consisting of poorly soluble hydrophobic free fatty acids (1). Polysorbate degradation is an industry-wide challenge both in biotherapeutics processing and formulation development. The risk of such degradation increases with higher cell densities and greater expression titers in bioprocessing, as well as with higher drug concentration in biologic formulations (2–3).
The main mechanisms for polysorbate degradation are oxidation and hydrolysis (1). Observations of temperature-dependent degradation that does not occur in placebo controls have led many experts to conclude that enzymatic hydrolysis is the major root cause of polysorbate degradation in biotherapeutics. Residual host-cell proteins (HCPs) such as lipases and esterases — including lipoprotein-associated phospholipase A2 (LPLA2), phospholipase A2, group XV (PLA2G15), lipoprotein lipase (LPL), carboxylesterase (CES), phospholipase A2, group VII (PLA2G7), lysosomal acid lipase (LIPA), and palmitoyl-protein thioesterase 1 (PPT1) — have been reported to show activity in PS-80 and PS-20 degradation (4–8).
Proteomics based on liquid chromatography with tandem mass spectrometry (LC-MS/MS) provides a means for simultaneous identification and quantification of HCP impurities and has emerged as an orthogonal assay for HCP characterization (9–11). Identification of individual HCPs is important to process development based on quality by design (QbD), with risk assessment to help us understand impacts on product quality and patient safety. Additionally, LC-MS/MS helps companies develop appropriate control strategies to mitigate the risk from specific residual HCPs and adapt the phase-appropriate HCP analytical testing strategy across product development and commercialization life cycles. Development of individual enzyme-linked immunosorbent assay (ELISA) kits or targeted proteomics-based approaches — e.g., multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM) — will benefit directly from LC-MS/MS proteomics identification work (5, 12).
Scientists using routine proteomics approaches for cell lysates must overcome two key analytical challenges in investigating the cause of PS-80 degradation. First, the dynamic range of HCPs present alongside a drug substance (DS) limits the detection sensitivity of HCP identification. Most reported proteomics-based approaches have detection limits at the sub-ppm level (with HCPs in abundance at six orders of magnitude over that of DS). Some lipases, such as LPLA2, can degrade polysorbate significantly when present at sub-ppm levels (5), thus going undetected by routine proteomics platforms.
Second, enzyme abundance cannot explain polysorbate degradation directly. Different high-risk HCPs have variable enzymatic activity — and thus, different effects related to polysorbate degradation. So determining enzyme activity is crucial to understanding the root cause of polysorbate degradation.
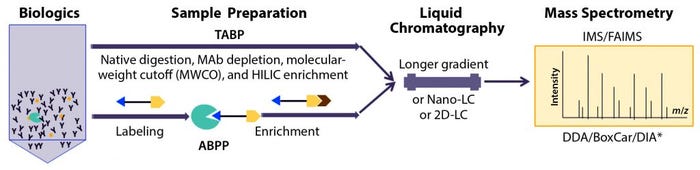
Figure 1: Recent advances in characterization of trace-level, high-risk host-cell proteins (HCPs) related to polysorbate degradation in biotherapeutic formulation by proteomics approaches based on liquid chromatography with tandem mass spectrometry (LC-MS/MS); method improvements in sample preparation for traditional abundance-based proteomics (TABP) and activity-based protein profiling (ABPP), LC, and MS.
* The BoxCar acquisition method for high-resolution MS-based proteomics combines acquisition of narrow m/z windows to enhance the mean ion-filling time compared with standard full scans. DDA-MS = data-dependent acquisition mass spectrometry; DIA-MS = data-independent acquisition MS; IMS = ion-mobility spectrometry; FAIMS = field-asymmetric ion-mobility spectrometry (FAIMS); HILIC = hydrophilic interaction liquid chromatography
Traditional Abundance-Based Proteomics
To drive the sensitivity of traditional abundance-based proteomics (TABP) approaches for scarce but high-risk HCPs, some researchers have focused on reducing the dynamic range of samples to improve assay sensitivity. Recent progress in this field covers the entire proteomics workflow, including sample preparation, LC separation, and MS analysis (Figure 1).
One widely used sample-preparation approach for HCP analysis is limited native digestion, developed by Huang and colleagues at Eli Lilly (10). Based on knowledge that antibodies are very stable and resistant to trypsin digestion in their native state, this approach maintains antibody structure and function while HCPs are digested. Compared with traditional sample-preparation methodologies for proteomics and peptide mapping experiments, the dynamic range of this method is expanded by one to two orders of magnitude for HCP detection by MS.
In a spike-in experiment, five recombinant Chinese hamster ovary (CHO) cell lipases or esterases — LPLA2, LAL, PPT1, phospholipase B-like 2 (PLBL2), and isoamyl acetate-hydrolyzing esterase 1 (LAH1) — were added into IgG1 or IgG4 monoclonal antibody (MAb) samples for evaluating the detection sensitivity of the native-digestion method (10). When the CHO proteins were spiked at ≥2 ppm, all five could be identified in both the IgG1- and IgG4-containing preparations with two or more unique peptides. Label-free absolute quantification is generally comparable with the spike-in amount, which is sufficient to support downstream process development. Using native digestion, the team was able to achieve a detection limit of 0.1 ppm by PRM quantitation (5).
Overall, the native digestion method is sensitive, fast, and robust, as confirmed by multiple publications (6, 13). It has become the benchmark for recent novel methods of HCP identification by proteomic means (13–16).
The field of HCP identification by proteomics has benefited significantly from accumulated knowledge from plasma and serum proteomics analyses with protein dynamic ranges known to exceed 10 orders of magnitude (17). A number of sample-preparation methods to remove high-abundance proteins or enrich low-abundance proteins has been used in HCP analysis to improve dynamic range and identification sensitivity (Figure 1): ProteoMiner kits from Bio-Rad Laboratories (15), drug-substance depletion with protein A (16), anti-HCP affinity chromatography (8), molecular weight cut-off (14), and hydrophilic- interaction liquid chromatography (HILIC) enrichment (18). For example, Chen et al. enriched low-abundance HCPs up to 1,000-fold using the ProteoMiner method (15). The team identified HCPs at a concentration ≥0.05 ppm after spiking in known amounts of HCPs. When applying that methodology to the study of HCPs present in the National Institute of Standards and Technology’s monoclonal antibody (NISTmAb) standard, the researchers confidently identified more than 500 HCPs.
In addition to sample preparation, improvements have been made to the sensitivity of HCP identification by LC-MS/MS, including lengthened chromatographic gradients, two-dimensional liquid chromatography (2D-LC), and nanoflow (“nano-LC”) systems, as listed in Figure 1 (13, 19). On the MS side, additional gas-phase separations with ion mobility include high-field, asymmetric-waveform, ion-mobility spectrometry (FAIMS) (16). Along with different MS acquisition technologies such as BoxCar multiplexed MS/MS and data-independent acquisition (DIA) (13), FAIMS has been used to improve HCP identification and quantification. These advanced LC-MS methods often get combined with improved sample preparation to achieve the best performance for HCP identification by proteomics.
For example, Nie et al. used a modified native digestion with ultralow trypsin concentration, long LC gradients, and BoxCar MS acquisition in a relatively simple and sensitive method for deep profiling of HCPs (13). The team identified more than 450 HCPs from the NISTmAb standard. In another study, Johnson et al. combined protein A depletion, native-digest conditions, and FAIMS for deep proteomic profiling of HCPs in MAbs (16). That team identified more than 600 HCPs (with at least two unique peptides) from the NISTmAb standard — representing the largest HCP data set yet identified from that standard. Note that five lipases/esterases and related HCPs were found.
Although sample-preparation methods (e.g., ProteoMiner kits and depletion of drug substance) do improve method sensitivity, they also increase the complexity of sample preparation and reduce sample-analysis throughput. Certain HCPs can get lost during depletion or enrichment. In addition to the effect on method robustness, these rigorous enrichment strategies also might decrease assay reproducibility while increasing variability in HCP identification and quantification.
Activity-Based Protein Profiling (ABPP)
Even with extensive fractionation, difficulties of dynamic range and ion suppression in MS from drug substances still exist. Moreover, there is a huge analytical gap between enzyme abundance and observed activity leading to polysorbate degradation. A method that could improve HCP dynamic range while detecting protein activity would drive the investigation of enzymatic degradation of polysorbate significantly.
Activity-based protein profiling is a chemical proteomic strategy for characterization of enzyme functional states in complex biological systems (20–21). ABPP offers an advantage over TABP techniques that rely on measuring abundance rather than functional activity of target proteins. Synthetic chemical probes — each comprising a reactive group, a linker, and an enrichment tag — are used to capture targeted active enzymes (Figure 1). The reactive group labels the active site of a mechanistically related enzyme molecule to form a covalent linkage (21). Then those covalently modified proteins are detected or purified based on the enrichment tags (e.g., biotin–avidin or alkyne–azide reactions) and identified with LC-MS (Figure 1).
ABPP first was developed to investigate enzymatic activity of serine hydrolases (SHs) (21) and cysteine proteases (22). Over the past decade, it has been adapted for activity detection of over a dozen enzyme classes that include proteases, kinases, phosphatases, glycosidases, and oxidoreductases (20–21). By studying enzymes with probes that label certain known enzyme classes specifically, researchers have been able to assign previously uncharacterized enzymes to new functional classes. That has been a key advantage of the activity-based proteomics approach.
SHs are one of the largest known enzyme classes, comprising more than 200 enzymes in humans (21). The class includes multiple lipases, esterases, thioesterases, amidases, and peptidases (21). The defining characteristic of the superfamily members is their active-site inclusion of a nucleophilic serine. A number of chemical probes have been developed for SHs (23). To confirm whether such active probes can block polysorbate degradation, my team’s recently published study demonstrated that polysorbate degradation can be blocked almost completely by chemical probes against SHs depending on timing and dosage (7). That has been confirmed by others using similar chemical probes or SHs inhibitors (24).
To optimize activity-based protein profiling for SH profiling in biologics process intermediates, several chemical probes against SHs were evaluated (6). A fluorescent protein (FP)–biotin probe was found to provide better enrichment for most SHs compared with the commercial FP–desthiobiotin probe (6), which was used in a recent ABPP application for discovery of CES in polysorbate degradation (7). All lipases or esterases that have been reported to cause polysorbate degradation have been identified by an ABPP approach: LPLA2 (5), LPL (4), CES (7), LIPA (8), and PPT1 (8). Binding specificities were confirmed by a competitive ABPP approach (6). In addition, PLA2G7 was identified and confirmed to contribute directly to polysorbate degradation for the first time (6).
Note that PLBL2 is the only enzyme with a lipase annotation that is not enriched by our activity-based protein profiling approach, which suggests that it most likely is neither an active lipase nor esterase (nor even an amidase or peptidase) in the SHs family because it is not enriched by the active chemical probes (6). An earlier study suggested that PLBL2 may be the root cause of PS-20 degradation to produce free fatty acids in a sulfatase drug product (25); however, the spiked-in amount of PLBL2 used for function confirmation was higher than that observed in drug products, and its purity from a non-CHO species was just about 90% (25). So the lipase activity observed from recombinant PLBL2 could come from contaminated lipases (26).
My team’s observation that PLBL2 was not an active lipase in the SH family is consistent with previously published data suggesting that
PLBL2 was proposed as an amidase or peptidase instead of a lipase (27)
purified recombinant CHO PLBL2 was reported to display no in vitro phospholipase activity with synthetic substrates (28)
a 20,000-fold higher PLBL2 concentration was required for similar hydrolysis relative to LPLA2 based on a fluorescence-based esterase activity assay (29)
according to the results of a genetic knock-out and immunodepletion experiment,
PLBL2 was not responsible for polysorbate degradation (26).
Overall, the impact of PLBL2 on polysorbate degradation at concentrations detected in biotherapeutics is questionable, although levels of PLBL2 need to be controlled because of immunogenicity risk (28). Direct confirmation by activity-based protein profiling that PLBL2 is not an active lipase is significant in the field of HCP analysis because it often is relatively abundant compared with lipases/esterases in bioprocess intermediates. PLBL2 has served as a surrogate HCP for demonstrating lipase clearance in process development. That testing strategy clearly needs to be revisited considering the accumulated evidence supporting that it most likely is not responsible for polysorbate degradation. More truly high-risk residual lipases/esterases have been identified by proteomics methods.
Using activity-based protein profiling to identify active enzymes for polysorbate degradation brings a number of challenges. First, although most lipases/esterases belong to the SH family (21), some lipases/esterases are members of other enzyme classes. For example, of the six types of PLA2 enzymes (30), four subtypes are considered to be serine proteases, and the other two use histidine as their active-site moiety (30). Whether host cells such as CHO express and secrete all types of PLA2 as HCPs remains a question to be answered. It is encouraging that a number of chemical probes/inhibitors against SHs have been demonstrated to mitigate polysorbate degradation almost completely (6, 7, 24).
Second, identifying active SHs with activity-based protein profiling does not confirm that those SHs have activities related to polysorbate degradation. Novel chemical probes yet could be designed specifically labeling lipase or esterase against polysorbate degradation.
Finally, the ABPP workflow should be optimized for detecting active lipases/esterases from biologic intermediates. Researchers now are working to determine how to load more starting material for enriching trace-level active enzymes while reducing nonspecific binding during sample preparation. Strategies for optimization of LC-MS conditions beyond sample preparation mentioned above also could be applied to improve the sensitivity of ABPP assays (Figure 1).
Future Perspective
Much progress has been made in the field to identify trace-level HCPs for polysorbate degradation in biotherapeutics using proteomics approaches based on LC-MS/MS. In addition to advancements in LC-MS optimization, sample preparation to improve dynamic range is key to success. Unbiased sample-preparation strategies have been used with TABP to identify global trace-level HCPs. Activity-based protein profiling using chemical probes specific for SHs has demonstrated unique potential to identify lipases or esterases for root-cause investigation of polysorbate degradation in biotherapeutics. That not only improves sample dynamic range, but it also provides additional enzyme activity information.
Both traditional abundance-based proteomics and activity-based protein profiling are relatively low-throughput options for sample testing during process characterization for lipase and esterase control of polysorbate degradation. Sensitive and high-throughput, plate-based esterase activity is an ideal tool for use in process development and characterization (29, 31). LC-MS/MS proteomics approaches including TABP and ABPP serve mainly as key tools for root-cause investigations that drive targeted process development related to polysorbate degradation. Sample-preparation strategies based on these proteomics approaches also can be used for development of middle-throughput, MS-based HCP quantification (e.g. MRM or PRM) with suitable internal standards. Trace-level, high-risk HCPs identified through optimized proteomics also will aid in development of individual HCP-specific immunoassays for potential high-throughput processes.
Acknowledgments
We are grateful for the fruitful discussions with our internal working groups on HCP-assay and polysorbate/lipase-control strategy.
References
1 Kerwin BA. Polysorbates 20 and 80 Used in the Formulation of Protein Biotherapeutics: Structure and Degradation Pathways. J. Pharm. Sci. 97(8) 2008: 2924–2935; https://doi.org/10.1002/jps.21190.
2 Tran B, et al. Investigating Interactions Between Phospholipase B-Like 2 and Antibodies During Protein A Chromatography. J. Chromatogr. A 1438, 2016: 31–38; https://doi.org/10.1016/j.chroma.2016.01.047.
3 Labrenz SR. Ester Hydrolysis of Polysorbate 80 in MAb Drug Product: Evidence in Support of the Hypothesized Risk After the Observation of Visible Particulate in MAb Formulations. J. Pharm. Sci. 103(8) 2014: 2268–2277; https://doi.org/10.1002/jps.24054.
4 Chiu J, et al. Knockout of a Difficult-to-Remove CHO Host Cell Protein, Lipoprotein Lipase, for Improved Polysorbate Stability in Monoclonal Antibody Formulations. Biotechnol. Bioeng. 114(5) 2017: 1006–1015; https://doi.org/10.1002/bit.26237.
5 Hall T, et al. Polysorbates 20 and 80 Degradation By Group XV Lysosomal Phospholipase A2 Isomer X1 in Monoclonal Antibody Formulations. J. Pharm. Sci. 105(5) 2016: 1633–1642; https://doi.org/10.1016/j.xphs.2016.02.022.
6 Li X, et al. Profiling Active Enzymes for Polysorbate Degradation in Biotherapeutics By Activity-Based Protein Profiling. Anal. Chem. 93(23) 2021: 8161–8169; https://doi.org/10.1021/acs.analchem.1c00042.
7 Zhang S, et al. Rapid Polysorbate 80 Degradation By Liver Carboxylesterase in a Monoclonal Antibody Formulated Drug Substance at Early Stage Development. J. Pharm. Sci. 109(11) 2020: 3300–3307; https://doi.org/10.1016/j.xphs.2020.07.018.
8 Graf T, et al. Identification and Characterization of Polysorbate-Degrading Enzymes in a Monoclonal Antibody Formulation. J. Pharm. Sci. 2 July 2021: https://doi.org/10.1016/j.xphs.2021.06.033.
9 Vanderlaan M, et al. Experience with Host Cell Protein Impurities in Biopharmaceuticals. Biotechnol. Prog. 34(4) 2018: 828–837; https://doi.org/10.1002/btpr.2640.
10 Huang L, et al. A Novel Sample Preparation for Shotgun Proteomics Characterization of HCPs in Antibodies. Anal. Chem. 89(10) 2017: 5436–5444; https://doi.org/10.1021/acs.analchem.7b00304.
11 Li X, et al. Identification and Characterization of a Residual Host Cell Protein Hexosaminidase B Associated with N-Glycan Degradation During the Stability Study of a Therapeutic Recombinant Monoclonal Antibody Product. Biotechnol. Prog. 21 January 2021: e3128; https://doi.org/10.1002/btpr.3128.
12 Gao X, et al. Targeted Host Cell Protein Quantification By LC-MRM Enables Biologics Processing and Product Characterization. Anal. Chem. 92(1) 2019: 1007–1015; https://doi.org/10.1021/acs.analchem.9b03952.
13 Nie S, et al. Simple and Sensitive Method for Deep Profiling of Host Cell Proteins in Therapeutic Antibodies By Combining Ultra-Low Trypsin Concentration Digestion, Long Chromatographic Gradients, and BoxCar Mass Spectrometry Acquisition. Anal. Chem. 93(10) 2021: 4383–4390; https://doi.org/10.1021/acs.analchem.0c03931.
14 Chen IH, et al. Improved Host Cell Protein Analysis in Monoclonal Antibody Products Through Molecular Weight Cutoff Enrichment. Anal. Chem. 92(5) 2020: 3751–3757; https://doi.org/10.1021/acs.analchem.9b05081.
15 Chen IH, Xiao H, Li N. Improved Host Cell Protein Analysis in Monoclonal Antibody Products Through ProteoMiner. Anal. Biochem. 610, 2020: 113972; https://doi.org/10.1016/j.ab.2020.113972.
16 Johnson RO, et al. Combination of FAIMS, Protein A Depletion, and Native Digest Conditions Enables Deep Proteomic Profiling of Host Cell Proteins in Monoclonal Antibodies. Anal. Chem. 92(15) 2020: 10478–10484; https://doi.org/10.1021/acs.analchem.0c01175.
17 Anderson NL, Anderson NG. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell Proteomics 1(11) 2002: 845–867; https://doi.org/10.1074/mcp.r200007-mcp200.
18 Wang Q, et al. Enhancing Host-Cell Protein Detection in Protein Therapeutics Using HILIC Enrichment and Proteomic Analysis. Anal. Chem. 92(15) 2020: 10327–10335; https://doi.org/10.1021/acs.analchem.0c00360.
19 Yang F, et al. A 2D LC-MS/MS Strategy for Reliable Detection of 10-ppm Level Residual Host Cell Proteins in Therapeutic Antibodies. Anal. Chem. 90(22) 2018: 13365–13372; https://doi.org/10.1021/acs.analchem.8b03044.
20 Sanman LE, Bogyo M. Activity-Based Profiling of Proteases. Annu. Rev. Biochem. 83, 2014: 249–273; https://doi.org/10.1146/annurev-biochem-060713-035352.
21 Long JZ, Cravatt BF. The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev. 111(10) 2011: 6022–6063; https://dx.doi.org/10.1021%2Fcr200075y.
22 Greenbaum D, et al. Epoxide Electrophiles As Activity-Dependent Cysteine Protease Profiling and Discovery Tools. Chem. Biol. 7(8) 2000: 569–581; https://doi.org/10.1016/S1074-5521(00)00014-4.
23 Wang C, et al. Discovery and Evaluation of New Activity-Based Probes for Serine Hydrolases. Chembiochem 20(17) 2019: 2212–2216; https://doi.org/10.1002/cbic.201900126.
24 Roy I, et al. Polysorbate Degradation and Particle Formation in a High Concentration MAb: Formulation Strategies to Minimize Effect of Enzymatic Polysorbate Degradation. J. Pharm. Sci. 30 May 2021; https://doi.org/10.1016/j.xphs.2021.05.012.
25 Dixit N, et al. Residual Host Cell Protein Promotes Polysorbate 20 Degradation in a Sulfatase Drug Product Leading to Free Fatty Acid Particles. J. Pharm. Sci. 105(5) 2016: 1657–1666; https://doi.org/10.1016/j.xphs.2016.02.029.
26 Zhang S, et al. Putative Phospholipase B-Like 2 Is Not Responsible for Polysorbate Degradation in Monoclonal Antibody Drug Products. J. Pharm. Sci. 109(9) 2020: 2710–2718; https://doi.org/10.1016/j.xphs.2020.05.028.
27 Repo H, et al. Is the Bovine Lysosomal Phospholipase B-Like Protein an Amidase? Proteins 82(2) 2014: 300–311; https://doi.org/10.1002/prot.24388.
28 Fischer SK, et al. Specific Immune Response to Phospholipase B-Like 2 Protein, a Host Cell Impurity in Lebrikizumab Clinical Material. AAPS J. 19(1) 2017: 254–263; https://doi.org/10.1208/s12248-016-9998-7.
29 Bhargava AC, et al. High-Throughput, Fluorescence-Based Esterase Activity Assay for Assessing Polysorbate Degradation Risk During Biopharmaceutical Development. Pharm. Res. 38(3) 2021: 397–413; https://doi.org/10.1007/s11095-021-03011-1.
30 Dennis EA, et al. Phospholipase A2 Enzymes: Physical Structure, Biological Function, Disease Implication, Chemical Inhibition, and Therapeutic Intervention. Chem. Rev. 111(10) 2011: 6130–6185; https://doi.org/10.1021/cr200085w.
31 Jahn M, et al. Measuring Lipolytic Activity to Support Process Improvements to Manage Lipase-Mediated Polysorbate Degradation. Pharm. Res. 37(6) 2020: 118; https://doi.org/10.1007/s11095-020-02812-0.
Xuanwen Li is a principal scientist, and Douglas D. Richardson is a distinguished scientist, both in analytical R&D mass spectrometry at Merck & Co., Inc., 2000 Galloping Hill Road, Kenilworth, NJ 07033; [email protected].
You May Also Like





