Residual Host-Cell DNA in Biopharmaceutical Products: 96-Well Plate-Based Extraction and Real-Time PCR Assay for Quantitative MeasurementResidual Host-Cell DNA in Biopharmaceutical Products: 96-Well Plate-Based Extraction and Real-Time PCR Assay for Quantitative Measurement
February 13, 2017
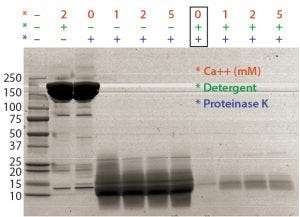
Figure 1: Optimal proteinase K digestion in the presence of detergent and/or calcium ion
Regulatory guidelines that cover the development of biopharmaceutical products require testing of host-cell deoxyribonucleic acid (DNA) impurities. Real-time polymerase chain reaction (PCR) has become a popular technology for DNA quantitation and monitoring of process impurities associated with biomanufacturing. One critical challenge associated with host-cell DNA impurity testing is that recombinant proteins (e.g., monoclonal antibodies, MAbs) and their corresponding buffer components often interfere with DNA quantitation in real-time PCR reactions (1, 2). Some sample types do not require a full extraction and can be quantitated by a real-time PCR directly. But because of the complexity of bioprocess sample matrices — from upstream to downstream processes — a full extraction typically must be executed.
We previously developed and validated a sodium iodide-based extraction method for removal of matrix interference from MAb therapeutic products (3). Although that method is simple, accurate, and precise for its intended use, it requires DNA extraction to be performed in single microfuge tubes that are repeatedly opened, closed, pipetted, and vortexed. The procedure was time consuming and labor intensive, and it became difficult for analysts to handle more than 30 sample tubes at a time, which ultimately limit the sample-testing throughput. Therefore, we propose a new extraction based on 96–deep-well plates to replace our tube-based method.
Materials and Methods
The “Materials” box lists all materials and instrumentation we used in this work. Diluted DNA was used as an assay standard and for spiking within the same assay day.
Materials and Instruments |
|---|
DNA EZ-Extraction kit (Ivy Fine Chemicals catalog #B48202) |
20 mg/mL proteinase K Invitrogen (Ivy Fine Chemicals catalog #25530-049) |
bovine serum albumin (BSA) and glycogen (Sigma-Aldrich catalog #7638 and #G8751, respectively) |
isopropanol (Fluka catalog #59304) |
2× PCR gene-expression master mix buffer (Applied Biosystems catalog #4304437) |
18 µM each of forward (5’-TGGAGAGATGGCTCGAGGTT-3’) and reverse (5’-TGGTTGCTGGGAATTGAACTC-3’) primers and 5 µM of a probe (6FAM-AGAGCACCAACTGCTGTTC CAGAGGTCC-MGB) based on carboxyfluorescein (FAM) dye and minor groove binder (MGB) (20× stock) (Applied Biosystems) |
CHO DNA standard purified from the CHO-K1 parental cell line using the QiaAMP DNA Micro kit (Qiagen #56304) and sonicated (scale 2 for one second) using a Misonix Sonicator 3000 instrument to 1–10 kb, which mimics fragmented genomic DNA in-process/intermediate biopharmaceutical samples — serially diluted from 100,000 pg/mL to 1 pg/mL in 10 mM Tris (pH 8.8) |
microcentrifuge and heating block (Barnstead Thermolyne type 28100 dry bath) |
Allegra X-15R plate centrifuge (Beckman Coulter) |
real-time type 7500 Fast PCR instrument (Applied Biosystems) |
Model 5336 Thermomixer R (Eppendorf) |
Nunc 96–deep-well plate and silicone cap (Thermo Scientific catalog #278743 and #276002, respectively) |
96-well insert (Millipore Sigma catalog #40-008) |
DNA Extraction: For a 96-well plate, the DNA extraction process must be significantly modified from our earlier tube method (3). We spiked 400 µL of samples at 5 mg/mL protein concentration with 40 µL of DNA standard. We spiked 400 µL of DNA standards at different DNA concentrations (1–100,000 pg/mL) with 40 µL of 50 mg/mL bovine serum albumin (BSA) protein to match the sample concentration at 5 mg/mL.
With samples, spiked samples, standards, and a blank control (10 mM Tris, pH 8.8), the 96–deep-well plate was incubated in a Thermomixer R incubator at 57 °C, 1,000 rpm for 40 min with 40 µL of a protein-digestion mixture containing kit detergent, 2M Tris, and 20 mg/mL proteinase K added to each well (3). We also added 1.1 mL of an ice-cold DNA precipitation mixture containing kit sodium iodide solution, 15 mg/mL glycogen, and isopropanol to each well (3). Tightly sealed with a Nunc silicone cap, the plate was manually inverted three times and left at room temperature for 20 minutes for DNA precipitation. Then we recovered DNA pellets by plate centrifugation at 2,700g for 20 minutes followed by a quick decant and drainage of residual supernatant on layers of paper towel (Kimberly-Clark WyPall or equivalent brand) for seven minutes (with a few changes of fresh paper towels required). Longer drainage time would not cause any risk of losing pellets.
We found no 70%-ethanol washing to be necessary for our applications, and it would have significantly reduced common assay variability from loss of DNA pellets. For evaporation of residual alcohol, we set the plate upright on a heating block at 55 °C for three minutes, then dissolved the DNA pellets in 400 µL water in the Thermomixer R incubator at 30 °C, 1,100 rpm for three minutes. Purified DNA could be analyzed on the same day (preferred) or kept frozen for a late testing.
The real-time PCR reaction was performed by an ABI 7500 Fast Real-Time PCR system similar to what is used in our tube method (3). We used multichannel pipetting to transfer the PCR reaction mixture containing a Taqman gene expression master mix and CHO 49 primers and probes, as well as to transfer purified DNA from the 96–deep-well plate to each well of the PCR plate. Then we sealed the latter, vortexed it on a thermomixer at 1,000 rpm for three minutes, and finally centrifuged it at 2,000 rpm for five minutes before quantitating by real-time PCR (3).
Results and Discussion
Assay Optimization: Most proteins aggregate at high temperatures, thereby interfering with DNA amplification if the aggregates are not digested out and removed efficiently before real-time PCR. Calcium ions can enhance proteinase K’s digestion by stabilizing the enzyme structure (4). So formulated bulk of a MAb was digested by proteinase K in combination with calcium and detergent. Detergent (not calcium) increased the efficiency of proteinase K digestion (Figure 1). Some of our intermediate samples and formulated bulks were in low pH and often precipitated during proteinase K digestion. To solve the precipitation issue and improve extraction efficiency, we added a 2M Tris base solution to the proteinase K digestion step.
Residual host-cell DNA often is fragmented by nucleases and shear forces in a bioreactor and/or during purification processes. Therefore, we matched the standards with samples by DNA size (<10 kb through sonication), protein concentration (5 mg/mL spiked BSA), and identical procedures in both extraction and real-time PCR. This experimental design minimized biases that may have affected quantitation of residual DNA level in samples with reference to the DNA standard used.
Plate Centrifugation and Decanting: With the significant expansion of our drug development pipeline, the previously developed DNA extraction in single-tube format did not meet our needs for high-throughput sample testing. So we explored the 96–deep-well plate format for its potential fit to our existing vortexing, heating, and centrifugation equipment, as well as its low protein and nucleic-acid binding surface.
Standard molecular biology practice involves recovering DNA from alcohol precipitation by a high centrifugation speed (e.g., >10,000 rpm). Plate centrifugation is designed to handle cells and can run only <3,000g force or 3,000–4,000 rpm. We found no report using low-speed centrifugation to recover fragmented DNA from alcohol precipitation efficiently and quantitatively at the pg/mL concentration range. Our validation data demonstrated that a low centrifugation speed of 2,700g force in combination with glycogen for coprecipitation could fully recover fragmented DNA quantitatively in the concentration range of 1–100,000 pg/mL using 96–deep-well plates. This information would allow many laboratories to perform DNA precipitation with 96–deep-well plates and bench-plate centrifuges, precluding the need for purchase of expensive high-speed instruments.
Contamination is a common challenge for PCR technology. This plate-extraction method involves sample DNA levels ranging 1–100,000 pg/mL. As such, well-to-well cross-contamination can be a major concern during decanting. We found filter plates from MilliporeSigma (without the filter, a customer special order) to fit tightly to the Nunc deep-well plate. So we inserted them into it before decanting as an attempt to eliminate cross-contamination from well to well.
Unfortunately, leaks still occurred, which led to cross-contamination and inconsistent results (data not shown). Moreover, during the mixing of samples with sodium iodide and alcohol for DNA precipitation, we found that the adhesive sealer detached from some wells, presenting another significant source of cross-contamination. Subsequent replacement with a Nunc silicone cap resulted in not only sealing the plate tightly and effectively, but also making the inefficient plate-insert unnecessary during the decanting step (see Assay Validation, below).
By comparison with our previous method using single microfuge tubes, this plate-based extraction method eliminates manual vortexing, reduced pipetting workload by 80%, cut down testing time from a full day to under half a day, and increased sample testing throughput by at least three-fold. The quality of data and analytical parameters of this method — e.g., limit of detection (LoD), limit of quantitation (LoQ), accuracy, and precision — remained as good as if not better than the earlier microcentrifuge tube method.
Although we also developed a plate-based technology using magnetic beads for DNA purification, it did not generate results equivalent to those described herein (data not shown), so is not described here.
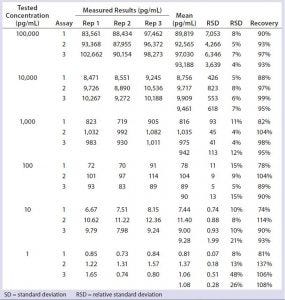
Table 1: Assay validation for measurement of residual Chinese hamster ovary (CHO) cell DNA in representative protein matrix (BSA at 5 mg/mL) by real-time polymerase chain reaction (PCR) — assay precision, limit of detection (LoD), accuracy, and limit of quantitation (LoQ)
Assay Validation: Compared with our validated tube method (3), the plate-based extraction significantly increases sample-testing throughput while significantly reducing time and workload related to vortexing and pipetting. With the improved assay performance at low centrifugation speed and elimination of well-to-well cross-contamination from a silicone cap sealer, we further validated the 96–deep-well DNA extraction and real-time PCR method for quantitation of both CHO and mouse residual DNA impurities according to our validation protocol and regulatory GMP and ICH guidelines (1). Our mouse DNA impurity assay was validated in an identical design to that described above, so it is not discussed herein.
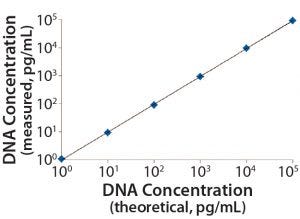
Figure 2: Linearity and range of the measured standards
DNA standards from 1 pg/mL to 100,000 pg/mL were spiked with 5 mg/mL of representative protein (BSA) solutions in triplicate, which we processed side-by-side with assay DNA standards through the identical DNA extraction and real-time PCR steps. Data from three independent assays run on three different days (Table 1, Figure 2) demonstrate a linear range from 1 pg/mL to 100,000 pg/mL, a linearity R2 at 1.00, LD at 1 pg/mL, and LoQ at 10 pg/mL. These results were similar to those obtained using the single-tube method (1, 3), indicating an increase in throughput for the optimized method, while maintaining the assays’ quality attributes.
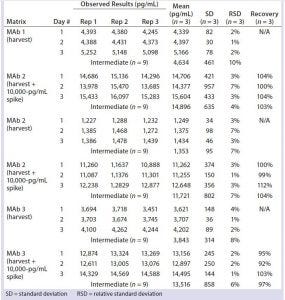
Table 2: Quantitation of residual DNA in harvest
We validated several product and intermediate matrices, including those from upstream bioreactor harvest, downstream formulated bulk, and column-cleaning samples for several MAbs in late stage clinical trials. We also evaluated the range of pH, salt, and detergent commonly used in our manufacture processes and final formulated bulk. And to determine assay robustness, we studied the range of incubation times, temperatures, and centrifugation speeds from the testing procedures.
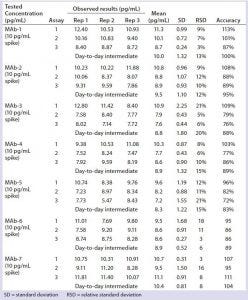
Table 3: Quantitation of residual DNA in formulated bulk
We diluted three late-stage bioreactor harvest samples 8,000-fold, spiked them with 10,000 pg/mL of DNA standard, and quantitated the results directly by real-time PCR without extraction or matrix interference (Table 2). Harvest matrix from three late-stage products was measured with an accuracy of 97–104% and a precision of 4–10% over three assays. We spiked formulated MAbs from three late-stage and four early stage products with or without 10 pg/mL of DNA standard and extracted and quantitated them by real-time PCR. No matrix interference was measured from the formulated bulk. Both precision (1–7%) and accuracy (92–112%) from each assay met our acceptance criteria (Table 3).
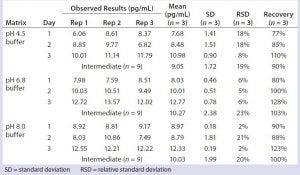
Table 4: Quantitation of residual DNA in representative column cleaning buffers
We prepared sodium acetate and Tris acetate buffers (common buffers in formulation and column cleaning) with salt concentrations ranging from 50–250 mM and pH values of 4.5–8.0. These buffers were spiked with 10 g/mL DNA standard, then extracted and quantitated by real-time PCR, and no matrix interference was observed (Table 4). This study allowed us to test similar but new product matrices without a need for matrix validation, thereby saving time and resources. We also tested sorbitol and sucrose (common formulation components) at 10%, and they showed no matrix interference in our validated qPCR assay (data not shown).
Matrix Interference from Phosphate Buffer: Phosphate buffer is commonly used in biological research as well as formulation of therapeutic MAb products. Thus we were surprised when our real-time PCR assay worked for all sample types but repeatedly failed with samples containing phosphate. The inhibition or interference from phosphate buffer to PCR amplification is poorly understood by biological researchers despite the fact that many biological samples contain phosphate salt. We found only one report on this inhibition of the PCR reaction and partial removal of it by overnight dialysis or sodium citrate treatment (5).
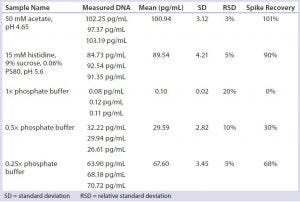
Table 5: Impact of phosphate buffer on qPCR results
In biological samples, phosphate salt often is present in the millimolar range, which is far higher than the phosphate group on each nucleotide (nM). The phosphate salt may compete nonspecifically with those nucleotide phosphate groups and would therefore interrupt the formation of phosphodiester bonds by Taq DNA polymerase during DNA amplification. Table 5 clearly demonstrates the real-time PCR inhibition by phosphate, but neither by other salt types nor by changes in pH.
For a quality control (QC) laboratory test method, it is neither practical nor efficient to remove phosphate salts from samples with overnight dialysis or sodium citrate treatment. Dilution of phosphate-containing samples in water, with further extraction for the removal of the phosphate’s matrix interference, was unsuccessful (data not shown). Neither did DNA pellet washing by 70% ethanol improve the DNA quantitation. It was unclear whether the poor DNA quantitation was caused by the leftover phosphate salt or simple DNA loss from the precipitation step.
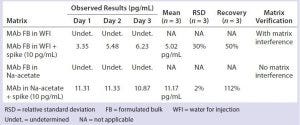
Table 6: Removal of phosphate salts by an alternative sample dilution in sodium acetate
We compared and processed a modified sample dilution in 50 mM sodium acetate (pH 4.5) side by side with sample dilution in water through DNA extraction and subsequent DNA quantitation by real-time PCR. The phosphate-containing samples diluted with sodium acetate buffer successfully demonstrated complete removal of phosphate’s matrix interference, whereas those samples diluted in water consistently failed the assay accuracy (as measured by % spike recovery) (Table 6).
This is the first report of which we are aware that not only proposes the possible mechanism of phosphate salt inhibition of the PCR reaction, but also provides a simple experimental approach to solve that matrix interference. Because of the popularity of PCR technology and common use of PBS buffer in biological research, we believe that our hypothesis and solution could help other scientists challenged by such phosphate interference.
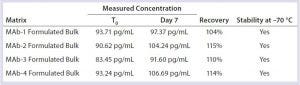
Table 7: Formulated-bulk stability with three freeze–thaw cycles
Sample Storage: DNA samples were reported stable at –20 °C for up to 16 months (6). It is important for sample submitters and testing laboratories to know the best storage condition(s) for residual DNA samples. For that purpose, we spiked representative final formulated bulks and column-cleaning buffers with 100 pg/mL of DNA, stored frozen (–70 °C) for seven days with one freeze–thaw cycle, two freeze–thaws, or three freeze–thaws during that week. We set the measured value from freshly spiked sample as time zero (T0) and 100%. Then we calculated the percentage recovered from stored samples from their measured concentrations in comparison with that of freshly spiked sample. We considered samples with a recovery of 70–130% to be stable.
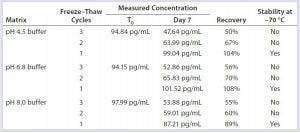
Table 8: Buffer stability
This study demonstrated that DNA was stable in formulated bulks when stored frozen at –70 °C. Up to three freeze–thaw cycles caused no significant DNA degradation in formulated bulks (Table 7). DNA was stable in column-cleaning buffers when stored frozen at –70 °C. However, repeated freezing and thawing caused significant DNA loss or degradation in column-cleaning buffers (Table 8). It is not clear why that DNA loss occurred in column-cleaning samples but not in formulated bulks. Therefore, we recommend –70 °C as a storage condition for all our DNA samples, although the number of allowable freeze–thaws for each given product can vary by product matrix.
Assay Automation: We scripted our validated qPCR assay procedures and automated them on a Tecan Freedom EVO platform. It prepared DNA standards, diluted samples, and transferred extracted DNA to PCR plates. The only steps that were not automated were centrifugation and decanting. Two consecutive valid assays demonstrated feasibility of the qPCR assay on the Tecan platform.
New and Improved
By comparison with the single-tube method we had developed for residual DNA quantitation (3), the validated 96-well plate method described herein maintained all the assay quality attributes (e.g., LoD, LoQ, precision, accuracy, linearity, and range) without the need for a washing step of DNA pellets. In addition, this new method increased testing throughput by at least threefold, reduced pipetting by 80%, eliminated vortexing during DNA extraction and PCR setup, and reduced testing time from 7–8 h down to 2–3 h.
Our validated, optimized residual DNA method provides the consistency and high efficiency required to recover a broad concentration range of fragmented DNA through low-speed centrifugation, and provides a new direction in high-throughput analysis of nucleic acids using common laboratory instruments. We also demonstrated automation feasibility with a TECAN platform. And we explored the mechanism of phosphate’s inhibition of PCR and identified a simple, effective, approach to remove phosphate from biological samples. This information raises awareness in — and provides a simple method to assist — the biological research community for solving a common, age-old issue of phosphate inhibition.
Acknowledgments
We thank Mehul Patel, Marianne Hayes, and Pat Niven for management support; and Krzysztof Mazuruk, Jeff Glenn, and Chi So for technical discussion and recommendations.
References
1 USP <1130> Nucleic Acid-Based Techniques: Approaches for Detecting Trace Nucleic Acids. United States Pharmacopeia: Rockville, MD, 2008.
2 Wang X, et al. Residual DNA Analysis in Biologics Development: Review of Measurement and Quantitation Technologies and Future Directions. Biotechnol. Bioeng. 109(2) 2012: 308–317.
3 Hu B, et al. Optimization and Validation of DNA Extraction and Real-Time PCR Assay for the Quantitative Measurement of Residual Host Cell DNA in Biopharmaceutical Products. J. Pharmaceut. Biomed. Anal. 88, 2014: 92–95.
4 Müller A, et al. Crystal Structure of Calcium-Free Proteinase K at 1.5-A Resolution. J. Biol. Chem. 269 (37) 1994: 23108–23111.
5 Johnson SR, et al. Alterations in Sample Preparation Increase Sensitivity of PCR Assay for Diagnosis of Chancroid. J. Clin. Microbiol. 33(4) 1995: 1036.
6 Jerome K, et al. Quantitative Stability of DNA After Extended Storage of Clinical Specimens as Determined by Real-Time PCR. J. Clin. Microbiol. 40(7) 2002: 2609–2611.
Further Reading
Cai H, et al. Development of a Quantitative PCR Assay for Residual Mouse DNA and Comparison of Four Sample Purification Methods for DNA Isolation. J. Pharmaceut. Biomed. Anal. 55, 2011: 71–77.
Zhang W, et al. Development and Qualification of a High Sensitivity, High Throughput Q-PCR Assay for Quantitation of Residual Host Cell DNA in Purification Process Intermediate and Drug Substance Samples. J. Pharmaceut. Biomed. Anal. 100, 2014: 145–149.
Corresponding author Bing Hu is principal scientist in CMC Analytical Services and Operations at Teva Biologics, West Chester, PA; 1-610-883-5588; [email protected]. Robert Peglow is a senior associate scientist, Matthew Miezeiewski is a scientist, David Colter is associate director, and Tam Soden is director of large-molecule analytical development, part of pharmaceutical development and manufacturing sciences at Janssen R&D, Johnson & Johnson in Malvern, PA. Shady Barsoum is a scientist in cell therapy at Janssen R&D, Johnson & Johnson in Spring House, PA.
You May Also Like





