Comparative Study of Single-Use and Reusable Fermentors: Production of Recombinant Proteins Through Bacterial FermentationComparative Study of Single-Use and Reusable Fermentors: Production of Recombinant Proteins Through Bacterial Fermentation
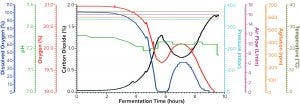
Figure 1a: Fermentation profile of protein A in the Sartorius SIP system at 10-L scale (dO2 in blue, pH in green, O2 in red, CO2 in black, pressure in cyan, air flow in magenta, agitation in orange, and temperature in olive); all controlling parameters in y-axes on the right were kept constant throughout growth. The y-axes on the left showed parameters mainly for monitoring, which changed through the fermentation process.
Single-use bioreactors have become widely accepted and well established for cell culture applications in the biopharmaceutical industry for over a decade (1). Abbott Diagnostics has moved into this technology already for commercial production of some biologic molecules. However, single-use systems (SUSs) are rarely available for microbial applications, mostly because of the technical challenge in designing cost-effective SUSs that can meet high oxygen transfer needs and remove excessive heat generated during fermentation. Thus, an important part of our biologics manufacturing — antigens from microbial fermentation — still relies on conventional stainless-steel stirred tanks.
Xcellerex XDR 50 single-use bioreactor
GE Life Sciences (www.gelifesciences.com)
Both Eppendorf and Sartorius Stedim Biotech have launched SUSs for fermentation, but theirs are more suitable for process development and scale-down experiments in design of experiment (DoE) studies (2, 3). Cellution Biotech has reported that its CELL-tainer single-use bioreactor could achieve kLa values comparable to those in traditional stirred vessels, thus obtaining microbial growth rates never before seen in SUSs (4). CELL-tainer 20-L bioreactors have ideal capacity and working volume to fit in our existing facility. However, this system is built on a different mixing mechanism than that used in stirred-tank systems.

HyPerforma S.U.F. 30 single-use fermentor, Thermo Fisher Scientific (www.thermofisher.com)
In recent years, both GE Healthcare (Xcellerex) and Thermo Fisher Scientific have launched single-use fermentors that mimic the principles of stainless-steel systems and reportedly match them in performance as well (5, 6). Here, we report on our evaluation of a GE XDR-50 SUS fermentor and a Thermo Scientific HyPerforma S.U.F. 30 SUS fermentor, comparing both with a Sartorius steam- or sterilization-in-place (SIP) fermentor at a working volume of 10 L for production of recombinant proteins.
Materials and Methods
Bacterial Strain: In this work, we used Escherichia coli BL21(DE3) containing a plasmid encoding the target proteins. Expression of the target protein was induced by either adding isopropyl beta-d-thiogalactopyranoside solution (IPTG) from Sigma when the culture optical density at 600 nm (OD600) reached a certain range, or by autoinduction using a medium following Studier’s method (7). Both approaches are used and reported widely for production of inducible proteins from cloned genes in E. coli. The choice of which induction method to use for each SUS was random.
Composition of Complex Medium: For all batch cultivations, we used a base complex medium consisting of
13.0 mL/L glycerol from Avantor (VWR catalog #5093-19)
24.0 g/L yeast extract from Becton Dickinson (#21730)
12.0 g/L tryptone from Becton Dickinson (#211701)
1.7 g/L monopotassium phosphate from Avantor (JT Baker #3248-01)
11.4 g/L dibasic potassium phosphate from Avantor (JT Baker #3250-05).
Hydrated medium was filtered through a 0.22-µm filter for the GE and Thermo fermentors and sterilized in place for the Sartorius SIP fermentor. We did not adjust pH before inoculation in any fermentor, but we measured it to be in the range of 7.2 ± 0.1 after medium preparation. For cultivations using autoinduction, we also added a premade autoinduction reagent to a final concentration of 1.6 g/L glucose (VWR #0188-2.5 KG from Avantor) and 6.6 g/L lactose (MilliporeSigma #17814-1kg). Before inoculation, we also added sterile ampicillin solution at 100.0 mg/L following fermentor setup.
Cultivation: A 100-mL seed culture was grown in a 250-mL sterile Corning disposable polycarbonate Erlenmeyer flask (or a 500-mL culture in a 1-L Erlenmeyer flask) until it reached an OD600 of 1.0 ± 0.1. We used that culture to inoculate the bioreactors for batch cultivation, having stored it in 4–8 °C until inoculation, which was within 24 hours of preparation.
Batch Cultivation: Batch cultivations for comparison ran in a 15-L Sartorius (formerly B. Braun ES15) stainless steel SIP fermentor with a working volume of 10 L. The unit has an aspect ratio of 3:1 and a bottom drive, its shaft equipped with 3 × 6-blade disk Rushton-type impellers. To prevent a vortexing effect, the vessel was mounted with four baffles.
The GE Xcellerex XDR-50 and Thermo Scientific HyPerforma S.U.F. 30 SUS systems used in this study had standard configurations recommended by their vendors, including sterile single-use vessels for mixing, exhaust venting, and air sparging with an available oxygen enrichment option. Each system has modules for monitoring and/or controlling agitation, air flow, temperature, dissolved oxygen (DO), and pH. Ports are available for aseptic connections to addition, sampling, and harvest. Each company’s website describes product features and system specifications (8, 9). We selected working volumes of 40 L (GE system) or 30 L (Thermo system), representing the upper end of the system capacities.

Table 1: Set points for controlling parameters in each fermentor system
All fermentations were controlled at 37 °C for their initial growth phase. Then, during the induction phase, they either were maintained at 37 °C for recombinant molecule A or were reduced to 25 °C for recombinant molecule B. We controlled pH to 7.2 (with a dead band of 0.1) by adding either a 1.6 M NaOH or a 0.8 M H2SO4 solution. Table 1 lists setpoints for all fermentation parameters. Those used with the Sartorius SIP fermentor were determined from earlier process development studies for each protein. Fermentation parameters for the SUSs were based on optimum operation ranges for each system as suggested by their vendors.
Analytical Methods: We took 10-mL samples at multiple timepoints during cultivation and determined the optical density of those samples at 600 nm. Each sample was centrifuged at 9,800 rpm for 15 minutes using a Beckman J2-21 centrifuge with a JA14 rotor. After decanting the supernatant, we calculated the weight of the pellet as wet-weight biomass growth. Then we used the cell paste to evaluate protein expression by sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) as follows.
Cell paste collected from the 10-mL samples as above was resuspended completely in BugBuster reagent from Novagen (catalog #71456-3), using 4 mL of reagent per gram of wet cell paste. We incubated the cell paste suspensions on a shaking platform set to 60 rpm for 15 minutes at room temperature, then centrifuged them at 16,000g for 15 minutes in room temperature before transferring the supernatant (representing the soluble fraction) to fresh tubes. We resuspended the pellets (representing the insoluble protein fraction) with 10 mL of phosphate buffered saline from Thermo Fisher Scientific (Gibco #10010031). Then we diluted both the soluble and insoluble fractions — three parts sample with one part 4× Laemmli sample buffer from Bio-Rad Laboratories (catalog #161-0791) — and placed them in boiling water for five minutes.
Boiled samples were loaded along with Precision Plus Dual Xtra Standard (Bio-Rad #161-0377) onto a Bio-Rad 4–20% Criterion Tris-HCl protein gel (#3450032). Gels ran at 150V in a Bio-Rad Power Pac HC system for 60–75 minutes, then were stained with Coomassie Brilliant Blue R250 dye (Bio-Rad #3450032) for an hour on a rocking shaker. We destained the gels in Coomassie Brilliant Blue R-250 destaining solution (Bio-Rad #161-0438) for 1.5 hours until the background became transparent. Destained gels soaked overnight in deionized water before they were imaged on a Bio-Rad GS800 imaging densitometer.
Results/Discussion
Fermentation for Production of Recombinant Protein A: Figure 1a shows the 10-L fermentation profile of pressure, agitation, air flow, temperature, pH, DO (%), and exhaust O2/CO2 concentration for recombinant molecule A from cultivation in the Sartorius SIP fermentor. Fermentation parameters that could be controlled during growth are shown on the right y-axis (pressure, airflow, agitation, and temperature); parameters we monitored during growth are shown on the left y-axis (DO, pH, and exhaust O2/CO2 concentrations). During this fermentation run, we kept the control parameters constant, as indicated by the flat lines in Figure 1a. Expression of recombinant molecule A was induced by IPTG when OD600 reached 5.46. After induction, the cultivation continued another four hours until OD600 reached 24.72 (Table 2). Then we stopped the fermentation and harvested the resulting cell paste.
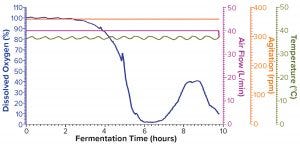
Figure 1b: Fermentation profile of protein A in the GE Xcellerex XDR-50 system at 40-L scale (dO2 in blue, air flow in magenta, agitation in orange, and temperature in olive)
Figure 1b shows the fermentation profile for cultivation of recombinant molecule A in a GE Xcellerex XDR-50 SUS fermentor. Again, parameters that could be controlled during growth are shown on the right y-axis, and the single parameter that we monitored during growth is shown on the left y-axis. This SUS does not offer the capability to control pressure; because of the nature of its disposable plastic vessel, the system can control only agitation, temperature, and air flow. In addition, the SUS does not have a built-in exhaust gas sensor or the capability to connect with a Blue-Sense off-gas analyzer for O2/CO2 concentration measurement. The system can monitor both DO and pH; however, we did not log the pH data during this study because of difficulties in operating the controller interface. As such, the left y-axis shows only the percentage of DO. Expression of recombinant molecule A was induced by IPTG when OD600 reached 9.23. After induction, the cultivation continued another four hours until OD600 reached 22.74 (Table 2), when we stopped the fermentation and harvested the cell paste.
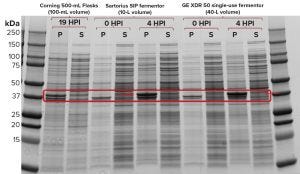
Figure 1c: Sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of recombinant molecule A samples from Sartorius SIP and GE Xcellerex XDR-50 fermentors; for reference, a sample was also loaded from production of the same protein in shaker flasks.
Figure 1c shows the results of our SDS-PAGE analysis of expressed recombinant proteins from cultivations in both systems as well as an overnight growth in a 500-mL shake flask (19 hours of growth after induction with 1 mM IPTG at an OD600 of 0.80). Evaluation at four hours postinduction (HPI) showed a strong protein band with similar intensity in the pellet fraction (P) for the SIP and SUS fermentors. Expression in the shaker flask was much lower, as expected, with no expressed protein observed in the soluble fraction (S).
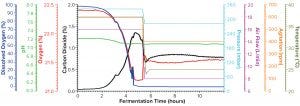
Figure 2a: Fermentation profile of protein B in the Sartorius SIP system at 10-L scale (dO2 in blue, pH in green, O2 in red, CO2 in black, pressure in cyan, air flow in magenta, agitation in orange, and temperature in olive); y-axes on the right were controlling parameters set higher for the first five hours of growth, then reduced to lower levels for the rest of fermentation (the protein-induction phase). The y-axes on the left are monitoring parameters, showing variation after changes in the control parameters and as fermentation progresses
Fermentation for Production of Recombinant Protein B: Figure 2a shows the 10-L fermentation profile of pressure, agitation, air flow, temperature, pH, DO%, and exhaust O2/CO2 concentration for recombinant molecule B from cultivation in the Sartorius SIP fermentor. Recombinant molecule B is expressed in the soluble fraction, and process development studies had demonstrated that slower growth during the induction phase enhanced protein expression (data not shown). Thus, five hours after inoculation, all controlled parameters were reduced: temperature from 37 to 25 °C, agitation from 600 to 400 RPM, air flow from 16 to 8 SLPM, and pressure from 345 to 270 mbar. At that point, the expression of recombinant molecule B through autoinduction was to initiate.
Figure 2a shows that change in controlled parameters on its right y-axis as a drop in the fermentation profile trace for those parameters at five hours. Because expression of recombinant molecule B started automatically with the depletion of glucose as cells switched to lactose for their carbon source, no other culture manipulations were required except for collection of samples to use in monitoring biomass and detecting protein expression.
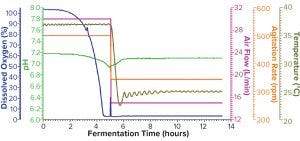
Figure 2b: Fermentation profile of protein B in the Thermo Scientific HyPerforma S.U.F. system at 30-L scale (dO2 in blue, pH in green, air flow in magenta, agitation in orange, and temperature in olive); same pattern of changes in controlling parameters (y-axes on the right) as in Figure 2a
Figure 2b shows the fermentation profile from cultivation in a Thermo Scientific HyPerforma SUS system run at its maximum capacity of 30 L. Again, controlled parameters are shown on the right y-axis, and monitored parameters are shown on the left y-axis. To shift protein expression into the soluble fraction, we reduced all three controlling parameters — agitation from 500 to 345 RPM, air flow from 30 to 15 L/min, and temperature from 37 to 25 °C — at hour five after inoculation. We stopped this fermentation at 13.4 hours when it reached the same level of biomass as had been achieved in the Sartorius SIP system (Table 2).
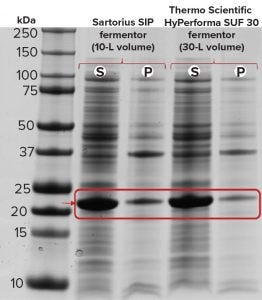
Figure 2c: SDS-PAGE of recombinant molecule B samples from the Sartorius SIP and Thermo Scientific HyPerforma S.U.F. 30 fermentors; S= soluble, P = pellet
Figure 2c shows our expression analysis of recombinant molecule B by SDS-PAGE from both the Sartorius SIP and Thermo Scientific HyPerforma SUS fermentors. Only endpoint samples are shown here because a zero-point sample is difficult to pull when autoinduction triggers the start of protein expression. Strong soluble expression occurred in both fermentation systems, with some expressed protein found in the insoluble pellet fraction.
Similar Results, So Far
Two recombinant molecules were expressed through IPTG or autoinduction in a Sartorius SIP fermentor and either a GE Xcellerex XDR-50 or a Thermo Scientific HyPerforma SUS at maximum batch size for that system. As Table 2 shows, both single-use fermentors reach similar optical densities as measured at 600 nm and similar cell paste weights to that achieved in the SIP fermentor. Expression levels also were comparably achieved in all fermentors for both recombinant molecules, as indicated by SDS-PAGE analysis (Figures 1c and 2c).

Table 2: Summary of fermentation outputs
Our limited study suggests that both SUSs could substitute for the fixed SIP system if larger quantities of either recombinant protein were needed for downstream applications. Of note is that both SUS fermentations were simple batch growths with low terminal densities, low oxygen demands, and low heat generation. How these SUS fermentors would perform with a high-density growth and feeding over an extended growth period remains to be tested.
Acknowledgments
We are grateful to GE Lifesciences for the GE Xcellerex XDR-50 demonstration unit. We thank Mark Gibson (senior process scientist from Lake County Operations) for valuable discussions on the single-use systems.
References
1 Shukla A, et al. Single‑Use Disposable Technologies for Biopharmaceutical Manufacturing. Trends Biotechnol. 31(3) 2013: 147– 154; doi:10.1016/j.tibtech.2012.10.004.
2 Application Note 293-I. Huether-Franken CM, et al. Scalability of Parallel E. coli Fermentations in BioBLU® f Single-Use Bioreactors. Eppendorf AG: Juelich, Germany, October 2013; https://www.eppendorf.com/product-media/doc/en/70274/DASGIP_Fermentors-Bioreactors_Application-Note_293_BioBLU-f_Scalability-Parallel-E-coli-Fermentations-BioBLU-f-Single-Bioreactors.pdf.
3 Pub. No. SBI1523-e160302. ambr® 250 High Throughput High Performance Single-Use Fully Automated Bioreactor System. Sartorius Stedim Biotech: Göttingen, Germany, 2016; https://www.sartorius.com/resource/blob/12004/8b7372f972446eda39c94f0300d244f2/broch-ambr-250-sbi1523-e-data.pdf.
4 Westbrook A, et al. Application of a Two-Dimensional Disposable Rocking Bioreactor to Bacterial Cultivation for Recombinant Protein Production. Biochem. Eng. J. 88, July 2014: 154–161; doi:10.1016/j.bej.2014.04.011.
5 Application Note 29-0564-39 AB. Microbial Fermentation in Single-Use Xcellerex™ XDR-50 MO Fermentor System. GE Healthcare: Uppsala, Sweden, February 2014; https://cdn.gelifesciences.com/dmm3bwsv3/AssetStream.aspx?mediaformatid=10061&destinationid=10016&assetid=17083.
6 Application Note CO29180. Brown J, et al. Scale-Up of Microbial Fermentation Using Recombinant E. coli in HyPerforma 30 L and 300 L Single-Use Fermentors. Thermo Fisher Scientific: San Jose, CA, 2014; https://www.thermofisher.com/content/dam/LifeTech/Documents/PDFs/CO29180-SUF-Launch-AppNotes-Scale-Up%20of%20Microbial-Global-FLR_V2.pdf.
7 Studier FW. Protein Production By Auto-Induction in High-Density Shaking Cultures. Prot. Expr. Purif. 41(1) 2005: 207–234.
8 Data Sheet: XDR Single-Use Bioreactors. Xcellerex, Inc.: Marlborough, MA, 2000; https://www.gelifesciences.co.jp/catalog/pdf/xdr-datasheet.pdf.
9 Catalog No. SUF0030.9001. HyPerforma™ Single-Use Fermentor Systems, 30L, Jacketed, AC Motor, with 2 Position Vent Filter Bracket. Thermo Fisher Scientific: San Jose, CA, 2019; https://www.thermofisher.com/order/catalog/product/SUF0030.9001.
Corresponding author Yongxue Ding is principal scientist, and Bill Zeck and Steven P. Allen are managers of biologics process design research and development at Abbott Diagnostics Division, Department 09NC, AP8A, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, IL 60064; 1-224-668-3629; [email protected].
To share this in PDF or professionally printed format, contact Jill Kaletha: jkaletha@mossbergco. com, 1-574-347-4211.
You May Also Like






