- Sponsored Content
- Bioreactors
- Process Monitoring and Controls
Dissolved Oxygen Control Tuning for Cell Culture ApplicationsDissolved Oxygen Control Tuning for Cell Culture Applications
Sponsored by Eppendorf
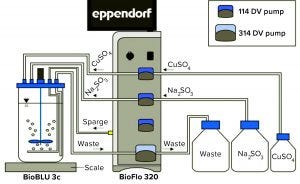
Figure 1: BioFlo 320 experimental setup
Proper tuning of dissolved oxygen (DO) controller proportional integral (PI) values is essential for optimal cell culture performance in a bioreactor. When DO-PI values are optimized, gas flows are smoothed, and foaming and cell stress are reduced. Traditionally, this tuning has been performed by using nitrogen gas to purge oxygen from a test solution, thus simulating oxygen demand. That method has several drawbacks, however. First, nitrogen gassing cannot simulate the high demands of high-density fermentation. Second, nitrogen competes with other gases, especially in single thermal mass-flow controller (TMFC) systems. The artificial gassing interference is not normally present in a cell culture. And third, nitrogen gassing cannot be used to test four-gas control mechanisms in which nitrogen is used to balance DO as part of the gassing strategy.
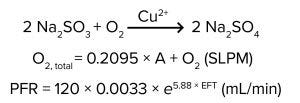
Equation 1: Reaction formula
Here, we describe a method of simulating DO demand using chemical oxygen scavengers such as sodium sulfite (Na2SO3), sodium metabisulfite (Na2S2O5), and sulfur dioxide (SO2). Oxygen scavengers have been used to deplete DO in a published protocol for oxygen transfer rate (OTR) measurements (1). In the presence of the right catalyst, oxygen scavengers quickly eliminate DO from solution. For example, with copper as a catalyst, sodium sulfite reacts with oxygen to form sodium sulfate (Equation 1).
Controlling the concentration of sodium sulfite and its feed rate into a bioreactor can simulate the oxygen demands of a cell culture. Iteratively, different PI values are used for each run to determine the most effective settings. The BioFlo 320 bioreactor system from Eppendorf offers a simple way to perform PI tuning simulations. Operators can use a built-in time-profile feature with integrated pumps to control the amount of a sodium sulfite solution added to a reactor over time. Below we describe this method for simulating culture oxygen demand for PI tuning, using Chinese hamster ovary (CHO) cells as an example, although it easily can be applied to other cell lines.
Materials and Methods
A BioFlo 320 system was used as the controller for this protocol. Simulations were performed in an Eppendorf BioBLU 3c single-use vessel with a macrosparger. All solutions were connected to the vessel using AdvantaPure pump grade silicone tubing from NewAge Industries. In lieu of culture medium, simulations used phosphate-buffered saline (PBS) from Thermo Fisher Scientific.
Sodium sulfite was prepared as a 0.5 M solution by first sparging nitrogen gas at 0.5 standard liters per minute (SLPM) into 5 L of deionized water for a minimum of half an hour to purge DO. Next, 315 g of anhydrous sodium sulfite (Thermo Fisher Scientific) was added quickly and carefully to the deionized water. The resulting solution was mixed vigorously with a stir bar until the sodium sulfite was completely dissolved. To inhibit oxidization, nitrogen was sparged continually into the solution at 0.1 SLPM for the remainder of the experiment.
The copper solution was prepared by dispensing 500 mL of deionized water and 25 g of copper (II) sulfate pentahydrate (Thermo Fisher Scientific) into a 500-mL glass bottle. The solution was mixed on a stir plate until all the copper (II) sulfate was dissolved. The bottle was wrapped in aluminum foil because the copper (II) sulfate is light sensitive.
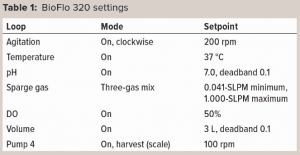
Experimental Setup: Figure 1 shows the experimental setup. The BioFlo 320 magnetic drive, thermometer, DO sensor, and heat blanket were connected to the BioBLU 3c bioreactor following instructions in the operator’s manual. A pH sensor was inserted into spare port 1, and the vessel was placed on a scale. After a zero-point and span calibration, the vessel was filled with 3 L of PBS. The BioFlo 320 loops were configured using parameters listed in Table 1.
 The copper (II) sulfate solution’s pump tubing was placed in one of the BioFlo 320 system’s integrated 114DV pump heads (Watson Marlow). Then 6 mL of copper (II) sulfate solution was pumped into the BioBLU 3c bioreactor to a concentration of 2 mL per 1 L of PBS. The Na2SO3 pump tubing was placed in another of the system’s integrated 114DV pump heads. The pump was primed, set to 0 mL/min, and turned on.
The copper (II) sulfate solution’s pump tubing was placed in one of the BioFlo 320 system’s integrated 114DV pump heads (Watson Marlow). Then 6 mL of copper (II) sulfate solution was pumped into the BioBLU 3c bioreactor to a concentration of 2 mL per 1 L of PBS. The Na2SO3 pump tubing was placed in another of the system’s integrated 114DV pump heads. The pump was primed, set to 0 mL/min, and turned on.
A waste jar was connected to the harvest line of the bioreactor through silicone tubing. That tubing was placed in the integrated 314DV pump head (pump 4). The pump was set to harvest mode to keep the vessel from overflowing as sodium sulfite was pumped in.
Time Profile Setup: Addition of sodium sulfite into the reaction vessel was controlled through the BioFlo 320 time profile. A simulated oxygen demand profile was created based on the increased oxygen demand over time observed during past CHO cultures. The simulated culture time was sped up by a factor of 10 so that a typical batch culture process of seven to 10 days was condensed into a one-day simulation. When the simulation was complete, data were exported from the BioFlo 320 system’s trend screen for analysis.
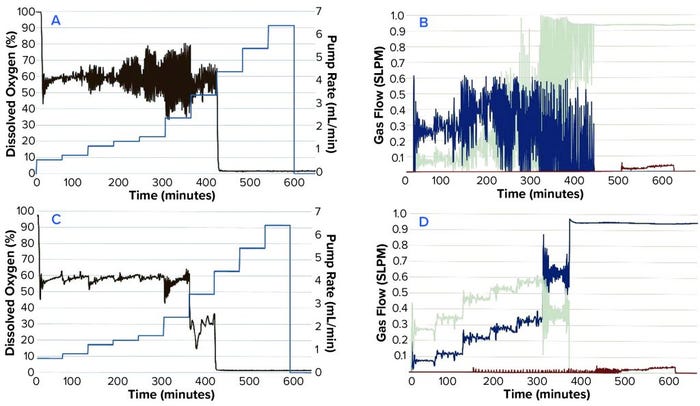
Figure 2: Comparing DO, N2SO3 pump speed, and gas flows in a time-profile simulation; (a) P = 5, I = 0.3; (b) P = 5, I = 0.3; (c) P = 2, I = 0.09; (d) P = 2, I = 0.09
Results and Discussion
We tested different settings around the default system PI values of P = 5 (0.7–5) and I = 0.3 (0.06–0.3). Figure 2A–D shows results from the time-profile simulation.
Step functions used in the time profile approximated the oxygen demands of an actual CHO culture. This method allowed us to simulate such demands sufficiently well to test different PI values efficiently. The basic PI tuning method does not require additional software or programming experience. In our CHO-cell batch culture example, values of P = 2 and I = 0.09 were found to be most effective. Those values reduced DO fluctuations around the setpoint and minimized fluctuations in gas flow.
To learn more about this method, visit https://www.eppendorf.com/bioprocess or contact us directly. Our experts will be happy to answer your questions.
Reference
1 How to Measure and Calculate OTR Using a New Brunswick Fermentor. Eppendorf, Inc.: Hamburg, Germany, 2013.
John Longsworth is a senior applications engineer, Nina Schillings is an intern, and Ma Sha is head of the applications laboratory at Eppendorf Bioprocess Center in Enfield, CT; [email protected].
You May Also Like





