Advancing In Silico Tools for Vaccine Development and Process ModelingAdvancing In Silico Tools for Vaccine Development and Process Modeling

The COVID-19 pandemic has demonstrated the importance of — and significant demand for — vaccines. However, vaccine development for large-scale manufacturing can be difficult and resource intensive because of the diversity and complexity of vaccine types that are needed. Developing a new vaccine typically takes more than a decade, with costs ranging from US$200 million to $500 million per successful program. That figure rises to $8 billion for epidemic vaccines (1). Vaccine programs also face a 90% risk of failure (2).
The Inno4Vac project intends to accelerate and minimize risks for vaccine development by tackling some major bottlenecks and creating innovative methods to identify the most promising vaccine candidates at early development stages. The Inno4Vac consortium is a public–private partnership established with financial support from the Innovative Medicines Initiative 2 (IMI-2), a joint undertaking of the European Union and the European Federation of Pharmaceutical Industries and Associations (EFPIA). With a budget of €38.5 million (~$41.2 million), Inno4Vac brings together more than 40 partners representing researchers from leading European academic institutions, innovative start-ups, and experienced industrial partners to address the consortium’s ambitious work plan.
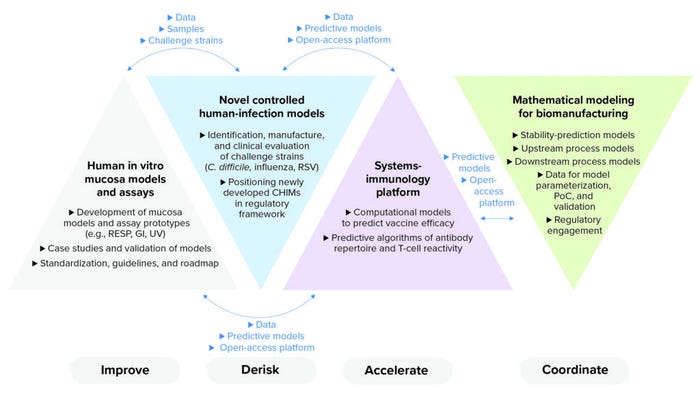
Figure 1: Inno4Vac project areas of action — summary and interaction (RESP = respiratory, GI = gastrointestinal, UV = urovaginal, RSV = respiratory syncytial virus, CHIMs = controlled human infection models, PoC = proof of concept).
The project will combine, optimize, and implement the latest advances in immunology, in vitro modeling, human-challenge models, and in silico modeling. That can be achieved by catalyzing collaboration among academic and industry researchers, biotechnology companies, public health agencies, and regulatory experts from across Europe. Results from the project will be shared with the broader scientific community through adherence to the principles of open science.
Negotiating Hurdles to Vaccine Development
Vaccine development is hindered by a number of scientific, economic, and infrastructural obstacles.Infectious agents cause some of the most pressing
global health problems, notably tuberculosis, malaria, and diarrheal and respiratory diseases. Lack of financial incentive means that few R&D investments are made to develop vaccines for such diseases. Effective and broadly acting vaccines for other infectious diseases — e.g., acquired immunodeficiency syndrome (AIDS) and influenza — have proven to be scientifically difficult to develop.
Inno4Vac will contribute to overcoming both financial and scientific barriers by developing new tools for making vaccine development and manufacturing faster and less expensive. A key component in the consortium’s efforts will be a thorough integration of in silico tools:
• an open-access and cloud-based platform to help predict vaccine efficiency
• both new and improved models for controlled human infection
• three-dimensional (3D) human-cell–based in vitro models that simulate infections at the mucosa and that more reliably predict immune protection
• an open-access, cloud-based, and modular one-stop computational platform for in silico modeling of vaccine biomanufacturing and stability testing.
Controlled Human-Infection Models
Traditionally, vaccine research has been empirical and thus time consuming and costly. Although the immunological mechanisms contributing to protective immunity are better understood today than they have been, large knowledge gaps remain in predicting the complex interplay between a pathogen and an infected person’s immune system. As consortium members, we will develop in vivo and in vitro human-infection models to increase our understanding of such relationships, complemented by in silico modeling and analytics based on artificial intelligence (AI) to tease out which factors help to determine the formation of protective immunity.
3D Human-Cell–Based In Vitro Models
During preclinical vaccine development, different animal models typically are used to confirm candidate safety and efficacy. Animal models are difficult to develop and have limited translational capacity. An alternative and complementary approach is to use a composition of different human cells and tissue scaffolds to simulate human organs — an organ-on-a-chip. Such technology enables simultaneous, high-throughput testing of multiple vaccine candidates and generates physiologically relevant data. Inno4Vac will develop and test next-generation 3D human in vitro models for gastrointestinal, respiratory, and urovaginal mucosae. Such systems will help to minimize use of animal models, reduce testing times, and improve our understanding of the protection mechanisms of the immune system.
Making Mathematical Models Work for the Vaccine Industry
Even though novel successful vaccine manufacturing platforms have been explored, approved, and implemented during the current pandemic, developing large-scale manufacturing processes remains difficult and resource intensive due to the wide variety and complexity of vaccine types. That breadth stands in contrast to the relatively narrow focus of areas such as monoclonal antibody (MAb) biomanufacturing. We are addressing the complexities of vaccine manufacturing by applying digital twins for stability and process modeling. Such approaches are enabling significant progress in expediting process development, control, and transfer between manufacturing sites.
Inno4Vac has developed a comprehensive approach to advancing biological and mathematical models of vaccine performance. Many models can simulate some aspects of vaccine development (e.g., vaccine design or prediction of individual immune reactions), but their relevance for biological immune responses remains variable. Current platforms also are not comprehensive enough to integrate development aspects together. For example, available tools can help to predict T-cell epitopes, but capabilities for predicting antibody binding sites are lacking. To address such gaps, experts from the Inno4Vac project will use biological data to refine tools that today predict responses of isolated immunological events and combine those into an integrated platform that provides relevant information about vaccine efficacy.
In terms of manufacturing areas, use of model-based approaches has accelerated in recent years, with process models supported by an increasing number of commercial offerings and with regulators often engaging with models for accelerated stability studies. But much remains to be done to increase their use and general acceptance. A significant factor limiting use of models in different aspects of drug development is regulatory concern over how such models might affect critical product parameters.
A major challenge for in silico tools is to demonstrate biological relevance. Today, that feat rests on limited high-quality input data. Although many excellent mathematical models are available for specific immunological aspects, few have been linked directly to biological observations of antibody formation or protection from disease. We expect that as different models demonstrate utility for biological analyses, they will be included naturally in all facets of vaccine development and evaluation.
Modeling Manufacturing Processes
Scale-up/-down and process transfer from one facility to another remain significant concerns in vaccine manufacturing. Developing in silico tools to optimize such processes can derisk them and reduce production costs. Stability models could help to ensure high drug-product quality.
Some advanced models for stability testing can help developers to determine vaccine product shelf life. To model entire vaccine manufacturing processes, much work remains to be done to integrate approaches for multiple unit operations and then use process and stability models to achieve model-based control. Consequently, regulatory acceptance of such models remains a long-term objective.
Thus, a key goal of Inno4Vac is to establish an open-source in silico simulation platform for guiding the design, scaling, operation, and transfer of vaccine manufacturing and stability testing. The data needed for model development will be acquired through public–private partnerships. By collecting such data, applying stability and process modeling, and obtaining validation from industry partners, we are expediting progress in activities such as process development, control, and transfer.
We will develop two open-source platforms, one to predict vaccine efficacy and another to optimize vaccine biomanufacturing. We will use biological data to refine today’s tools for predicting isolated immunological events, then combine those into an integrated platform that can help to predict vaccine efficacy. Current trends surrounding digitization and manufacturing in the biopharmaceutical sector indicate a receptiveness to the goals of our project.
Each Inno4Vac subgroup is working to identify relevant data sets and/or establish predictive models. Project partners already have established a sample manufacturing process based on data for a protein subunit vaccine expressed by Escherichia coli cells. That process will be used to build models.
Because we seek to develop in silico models that can be implemented in different aspects of vaccine design and manufacturing, regulatory considerations are critical. Dialogue with regulatory agencies has begun to ensure that developed models won’t be theoretical, but rather applied to accelerate vaccine development.
Understanding Protective Immunity
As the consortium increases its understanding of the principles underlying formation of protective immune responses, our in silico models can be improved correspondingly. A key factor in developing a predictive model lies at the intersection of AI, “big data,” and biological experimentation. Developing in vitro assays and human-infection models will help us understand that relationship. Such work will be complemented by in silico modeling of the immune system and applications of AI to identify mechanisms for the formation of protective immunity. AI could be particularly relevant for identifying key patterns in a complex immunological picture so that we can better understand the governing features of vaccine efficacy.
References
1 Plotkin S, et al. The Complexity and Cost of Vaccine Manufacturing — An Overview. Vaccine 35(33) 2017: 4064–4071; https://doi.org/10.1016/j.vaccine.2017.06.003.
2 Gouglas D, et al. Estimating the Cost of Vaccine Development Against Epidemic Infectious Diseases: A Cost Minimisation Study. The Lancet: Global Health 6(12) 2018: e1386–e1396; https://doi.org/10.1016/S2214-109X(18)30346-2.
Acknowledgments
This project has received funding from the Innovative Medicines Initiative (IMI) 2 Joint Undertaking under grant agreement 101007799. This Joint Undertaking receives support from the European Union Horizon 2020 research and innovation program and the European Federation of Pharmaceutical Industries and Associations (EFPIA).
Corresponding author Irina Meln is senior innovation manager at the European Vaccine Initiative (EVI), UniversitätsKlinikum Heidelberg, Voßstraße 2, Geb. 4040, 69115 Heidelberg, Germany; [email protected]. Gunnveig Grødeland is senior scientist and research group leader at Oslo University Hospital. Daniel G. Bracewell is professor of bioprocess analysis at the University College London (UCL) Department of Biochemical Engineering. Cécile van Els is a subject matter expert in immunology of infectious diseases and vaccines at the National Institute for Public Health and the Environment (RIVM) in The Netherlands. Meta Roestenberg is a professor in vaccinology and clinical head of the Controlled Human Infection Center at the Leiden University Medical Center in The Netherlands. Kimberly Veenstra is a project manager, and Ole Olesen is executive director of EVI. This communication reflects the authors’ view, and neither the Innovative Medicines Initiative nor the European Union, European Federation of Pharmaceutical Industries and Associations, or any associated partners are responsible for any use that may be made of the information contained herein.
You May Also Like





