Evaluating Adsorptive Filtration As a Unit Operation for Virus RemovalEvaluating Adsorptive Filtration As a Unit Operation for Virus Removal
Xenotropic murine leukemia virus (XMuLV), one of the model viruses used in this study. WIKIMEDIA COMMONS (HTTPS://COMMONS.WIKIMEDIA.ORG)
Most recombinant monoclonal antibodies (MAbs) are produced by mammalian cells. Because biopharmaceuticals derived from mammalian tissue culture carry the risk of adventitious virus contamination, regulatory agencies expect risk-mitigation strategies to include validation of purification unit operations for their ability to clear viruses (1). Guidelines from the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) describe how to prove viral clearance in downstream purification processes using an orthogonal approach (2). Viral log10 reduction values (LRVs) are determined for orthogonal steps and combined to calculate the cumulative log viral reduction for an entire process (3, 4). Regulatory agencies typically expect drug sponsors to be able to describe their mechanisms of viral clearance in detail (4).
Downstream processing of MAbs commonly starts with protein A capture chromatography. Because most MAbs are eluted at low pH (5), a viral inactivation step follows with incubation between pH 3.0 and 4.0 (4). Low-pH treatment and neutralization leads to precipitation of impurities, so a filtration step is required for particle removal. Depth filters are commonly used to combine removal of precipitates with adsorptive binding of remaining soluble contaminants (6). In addition to removing host cell proteins (HCPs) and DNA (7), viral clearance of >4 LRV has been reported for positively charged depth filters (8–11).
In a previous virus clearance study, we demonstrated the capability of charged 3M Zeta Plus depth filters and 3M Emphaze AEX Hybrid Purifier devices as adsorptive unit operations for virus clearance (8). At pH 5.5, removal of minute virus of mice (MVM) with LRVs of 4.6–7.2 were observed for both at 220 L/m2 throughput. Removal of MVM at pH 7.0 decreased significantly to LRVs of 2.4–2.7. Clearance of the larger xenotropic murine leukemia virus-related model virus (X-MuLV) also was determined at common process conditions (pH 5.5 and pH 7.0).
Additionally, we tested pH 7.0 at high conductivity (1 mol/L NaCl) to suppress ionic interactions. Under low-salt conditions at pH 5.5 and 7.0, X-MuLV clearance ranged from 5.0 to 5.8 LRV for all tested filter types. By comparison, adding NaCl substantially lowered clearances to 1.1 LRV (Emphaze) and 2.8 LRV (Zeta Plus). The difference in LRVs between low- and high-salt conditions were attributed to anionic adsorption. By arithmetric subtraction, the Emphaze device showed X-MuLV clearance relying on ionic interactions of 4.0–4.7 LRV, whereas Zeta Plus depth filters delivered 2–3 LRV. Residual X-MuLV clearance obtained under high-salt conditions (1.1–2.8 LRV) may be attributed to hydrophobic interactions or mechanical retention.
Based on those results, we have investigated the retention mechanisms of charged depth filters in more detail, again using the small nonenveloped model virus MVM (20–25 nm) and the larger enveloped virus X-MuLV (80–100 nm). To differentiate the mode of virus removal related to ionic, hydrophobic, and mechanical retention, we performed filtration runs at low conductivity, high conductivity, high propylene glycol (PG) concentration, and high conductivity with PG. Additionally, we compared the results of our virus spiking study with HCP removal levels determined at those four experimental conditions. The results provide a detailed understanding of mechanisms involved in virus retention by charged depth filters.

Table 1: Overview of the used filter types and process parameters
Materials and Methods
Materials and Instruments: For our adsorptive depth filtration studies, we used ÄKTAexplorer 10 systems with Unicorn software from GE Healthcare. All buffers were 0.2-µm filtered before use. Table 1 lists the adsorptive filters with anion-exchange functionality.

Table 2: Virus models used for the virus clearance study
Load Material and Virus Assay: Virus-spiking experiments were performed at Charles River Laboratories (CRL) in Cologne, Germany, using X-MuLV and MVM as model viruses. Table 2 provides more detailed information. The load material for our study was prepared by processing a MAb-containing harvest. Culture harvest was clarified with a Zeta Plus 60SP02A device and 0.2-µm filtered before protein A affinity chromatography eluted with a phosphate–citrate–Tris buffer. The eluate was adjusted to pH 5.5 and diluted to MAb concentrations of 10 g/L and 15 g/L. Samples were 0.2-µm filtered and stored at –70 °C until the spiking study could be performed. Before the adsorptive depth filtration runs, load samples were thawed and adjusted to our desired loading conditions regarding NaCl and PG concentration (Table 3). Those adjustments were made with buffer stock solutions containing 4 mol/L NaCl and 80% PG, respectively, leading to a final MAb concentration of 7.5 g/L for all sample loads.

Table 3: Experiments were performed in duplicate except for run #1 condition already tested in a previous study).
To exclude any influence of test items on cell growth or virus replication, CRL also performed cytotoxicity and viral interference assays. Providing the virus stock solutions, the test facility further analyzed samples with 50% tissue culture infective dose (TCID50) assays. The detection limit of such assays depends on the volume of sample incubated with indicator cells. Those cells were cultivated for a specific incubation period and inspected microscopically for virus-induced changes in cell morphology.
Experimental Methods: Virus-clearance experiments on adsorptive depth filters used four different buffer conditions (Table 3). Virus-reduction values for X-MuLV and MVM were determined using phosphate–citrate–Tris buffer (pH 5.5) but without adding NaCl (“pH 5.5 low salt”), representing an appropriate condition for further processing in a MAb purification process. Second, viral reduction was determined using a phosphate–citrate–Tris buffer at pH 5.5 with 1 mol/L NaCl (“pH 5.5 high salt”) to minimize ionic interactions between viruses and the adsorptive filters. Next, experiments using a phosphate–citrate–Tris buffer at pH 5.5 with 20% PG (“pH 5.5 high PG”) were carried out to minimize hydrophobic interactions. Finally, viral reduction was determined using a phosphate–citrate–Tris buffer at pH 5.5 with 1 mol/L NaCl and 20% PG (“pH 5.5 high salt, high PG”) to minimize ionic and hydrophobic interactions for assessing a possible mechanical virus-retention effect with these filters.
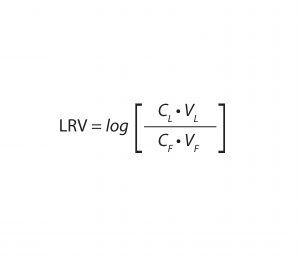
Equation 1: Calculation of virus log10 reduction value (LRV), with CL = virus concentration of the load, VL = volume of the load, CF = virus concentration of the filtrate, and VF = volume of the filtrate
Filtration runs were performed in parallel with two chromatography systems. CRL spiked 180 mL of protein-A–purified and conditioned MAb solutions with 5% (v/v) of ultracentrifuged and prefiltered virus stock solution (0.45 µm for X-MuLV, 0.1 µm for MVM). After mixing, the laboratory immediately analyzed a sample from each spiked pool for virus titer (“load sample”). For X-MuLV, CRL stored a second load sample until the end of the filtration (“hold sample”). MVM is physicochemically resistant, so no hold sample was analyzed for it (10).
Analysts loaded 220 L/m2 (84 mL) of starting material at 158 L/m2/h (1 mL/min) onto equilibrated adsorptive depth filters, then flushed them with 63 L/m2 (24 mL) equilibration buffer. Finally, they analyzed virus titers of total filtrates (flow-through and flush, the “filtrate sample”) and hold samples. Virus log10 reduction values (LRV) were calculated as described in Equation 1.
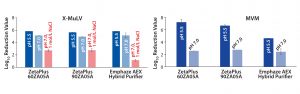
Figure 1: Log10 reduction values (LRVs) for (A) X-MuLV and (B) MVM at low- and high-salt conditions with three filter devices (8). Difference in X-MuLV clearance between low- and high-salt concentration (red arrows) is attributed to electrostatic adsorption. Residual LRV at 1 mol/L NaCl might be related to hydrophobic or mechanical retention. Because MVM is much smaller than the filters’ nominal pore size, mechanical retention is excluded, and runs at high-salt conditions were not performed.
Results and Discussion
Cytotoxicity and viral interference tests performed by CRL determined necessary dilutions that would not influence cell growth or virus replication. A recovery assay showed that all X-MuLV–spiked starting materials could be held at room temperature for up to four hours without significant loss of virus titer (data not shown).
Removal of X-MuLV: Clearance of X-MuLV by a Zeta Plus 90ZB05A depth filter showed comparable log10 reduction values (LRVs) for all four tested conditions (Figure 2, top). Regardless whether NaCl, PG, or both were added at pH 5.5, the determined LRVs ranged from 1.8 to 3.0. Because NaCl and PG did not influence virus clearance, the results indicated that removal of the remaining 2 LRV could be attributed to size exclusion. Previously stated importance of hydrophobic interactions in X-MuLV retention by Zeta Plus depth filters (12) could not be confirmed in our study. Compared with promising results obtained in our first study — which delivered X-MuLV clearance of 6 LRV at pH 5.5 under lowsalt conditions (Figure 1A) — X-MuLV clearance was significantly lower (2.2 LRV) in the present study. Because the results under high-salt conditions are comparable (2.8 log, 1.8 log), our proposed hypothesis of mechanical retention by size is supported.
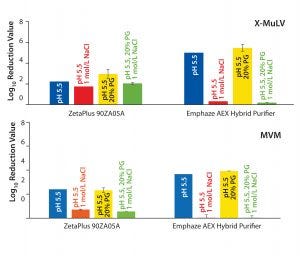
Figure 2: Log10 reduction values (LRVs) for (top) X-MuLV and (bottom) MVM at low-salt, high-salt, high-PG, and high-salt + high-PG conditions with two filter devices
Several factors could alter the effectiveness of virus clearance by depth filtration. Lower clearance determined under process conditions (pH 5.5) could relate to the three parameters varied from the first to the second study: filter media lot, MAb molecule, and total MAb load (which was 825 g/m2 in the first study and 1,650 g/m2 in the second). The higher total MAb load could have led to hydrodynamic-radius–based shielding of binding sites on the filter media, decreasing the number of available binding sites for viruses (9). Because filtration runs in both studies were performed in duplicate using the same filter lots, some lot-to-lot variation between the two lots used in these studies is conceivable. Zeta Plus depth filters are made of natural ingredients (diatomaceous earth and cellulose), so some variation between lots is possible. That could cause differences in pore structure inside the filter media and ultimately influence mechanical entrapment (13).
Evaluated in separate experiments, Zeta Plus 90ZB05A depth filters are composed of two different filter layers (Table 1), with the upstream 30ZB layer having pore sizes significantly larger (0.5–2.0 µm) than the X-MuLV model virus (80–100 nm). Because size-related retention thus can be excluded, we considered virus clearance by that single filter layer to be easier to validate than that of the whole filter. So we had the laboratory fractionate the filtrate and flush material from its 30ZB runs into three fractions (virus-removal capacity of a single 30ZB layer could be lower than that of another because of the half thickness of the filter media).
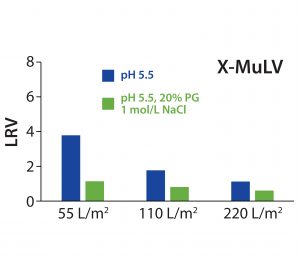
Figure 3: Log10 reduction values (LRVs) for X-MuLV at different filter loadings for low-salt or high-salt–high-PG conditions with 30ZB filter layer (upstream layer of the Zeta Plus 90ZB05A device)
X-MuLV clearance of the first fraction was high after loading of 55 L/m² at pH 5.5, with a calculated LRV of 3.8 (Figure 3). It significantly decreased at a higher loading of 110 L/m², and further at a complete loading of 220 L/m2, down to a 1.1 LRV. That decrease in virus removal can be explained by exhausting the electrostatic adsorptive capacity of the depth-filter media. With simultaneously high salt and PG concentrations, consistently low LRVs (0.6–1.1) confirmed that the retention was not based on a size-exclusion effect. Nevertheless, the loading capacity for higher viral clearance is limited to 55 L/m2 (corresponding to a MAb load of 412.5 g/m2), but that could be compensated in practice with increased filter area. However, it would be necessary to prove whether viral clearance capacity remains as high as measured when a flush step is carried out after a loading of 55 L/m2.
By contrast, results from the synthetic Emphaze device confirmed the X-MuLV results in our first study (Figure 1A). Again, we determined virus removal of 5 LRV under low-salt conditions (pH 5.5), whereas adding NaCl suppressed ionic interactions and reduced LRV to 0.32. That observation again was supported by results from the high-PG (LRV 5.5) and the high-salt–high-PG (LRV 0.2) conditions. Because only the addition of NaCl suppressed virus removal, we believe that the retention mechanism is based predominantly on ionic adsorption, with no hydrophobic effects. We expected that based on the construction of the device, two-thirds of its functionalized nonwoven mass holding the cationic polymer was responsible for an electrostatic adsorption.
Removal of MVM: Because the nominal pore sizes of Zeta Plus 90ZB05A and Emphaze AEX Hybrid Purifier filters are significantly larger (>0.2 µm) than MVM (20–25 nm), we did not expect virus removal by size exclusion. Nevertheless, we tested all four conditions to determine whether the retention is related to ionic and/or hydrophobic interactions. 90ZB05A results showed MVM clearance of 2.3–2.4 LRV for pH 5.5 + low salt and pH 5.5 with 20% PG conditions (Figure 2, bottom). Thus, we can assume that hydrophobic interactions are not relevant for MVM retention. By contrast, virus clearance significantly decreased for both conditions spiked with NaCl to LRV <0.7, a level considered to be insignificant. Thus, the determined LRVs under low-salt conditions (2.3–2.4) are associated with ionic interactions. Moreover, a size-exclusion effect could be excluded for MVM because no substantial removal of viruses could be observed when electrostatic adsorption was suppressed by NaCl.
So the 90ZB05A filter removed MVM in the range of 1.5–2.0 logs based on anionic adsorption. Comparing these results with the high LRV of 6.7 at pH 5.5 determined in our previous study (Figure 1B), we see that the clearance is substantially lower herein. This finding is in accordance with results obtained for X-MuLV above. Again, possible reasons include higher total MAb load and lot-to-lot variability among depth filters.
An Emphaze AEX Hybrid Purifier device delivered MVM clearance of 3.7–3.9 LRV at pH 5.5 under low-salt conditions (Figure 2, bottom). By contrast, with a 1-mol/L NaCl spike, LRVs were insignificant (LRV <0.03). Adding PG to suppress hydrophobic effects made no difference. These results confirm that retention of MVM on this depth filter also is based predominantly on electrostatic adsorption, as already shown for X-MuLV above. Clearances were similar to the result of our first study with a determined LRV of 4.6 at pH 5.5 (Figure 1B). As a result, higher MAb loads and different filter lots appeared to have no significant influence on the device’s virus-removal capacity.
Removal of Host Cell Proteins (HCPs): To broaden our knowledge about retention mechanisms for the conventional and synthetic depth filters, we performed additional tests to determine HCP clearance for all conditions we tested.
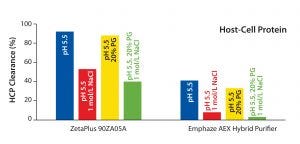
Figure 4: Host cell protein (HCP) clearance at low-salt, high-salt, high-PG, and high salt + high PG conditions for MAb 2 (CHO HCP Kit #F550, third generation, from Cygnus Technologies)
We found it interestingly that HCP clearance was influenced by hydrophobic interactions in neither filter. HCP removal at pH 5.5 was comparable with that at pH 5.5 using 20% PG (Figure 4). That result is in accordance with results of the virus study above (Figure 2). Under conditions including 1 mol/L NaCl, HCP clearance by with the 90ZB05A filter decreased by about 50%, whereas removal of HCP with the synthetic device decreased by about 85% (Figure 4). Thus, HCP clearance of the latter fully relies on anionic adsorption, whereas HCP removal capability of the conventional filter involves multiple retention modes.
Even though HCPs represent a mix of proteins of different sizes and electrical charges, these results clearly support our proposed retention mechanisms for both model viruses on the two filters.
Electrostatic Adsorption
To improve our understanding of the mechanisms of virus retention on both Zeta Plus and Emphaze depth filters, we had a contract laboratory perform virus-spiking runs under four different conditions. We chose those feed stream conditions — pH 5.5 with low conductivity, pH 5.5 with high conductivity, pH 5.5 with high PG, and pH 5.5 with high conductivity and high PG concentration — to evaluate which interactions contributed to viral clearance: ionic, hydrophobic, or mechanical. In addition, we investigated mechanical retention of the enveloped X-MuLV virus through experiments using the upstream filter layer of the Zeta Plus 90ZB05A filter with its higher nominal pore size (30ZB) as a reference.
By contrast with previously published suggestions concerning the binding mechanisms on conventional depth filters (9, 12, 14, 15) we found that hydrophobic effects are not important for the retention of MVM and X-MuLV on the Zeta Plus 90ZB05A depth filter because the use of PG did not influence determined LRVs. Current results showed only minor ionic retention, whereas a significant virus clearance based on electrostatic adsorption was reported in our previous study (8). This indicated that the virus clearance capability of the depth filter is subjected to variations. Not even Zeta Plus filters are completely specified, although they are characterized by defined specifications with respect to the amount of accessible charge, the uniformity of the media, and/or the charge distribution at submicron level (7). Therefore, it is possible that lot-to-lot variations or a strong dependency on the MAb loading are dominantly responsible for these observations. The determined LRVs at high conductivity and high PG indicated a residual retention based on the physical entrapment of X-MuLV (2 LRV). As presumed, no size-exclusion effect was determined for the smaller nonenveloped model virus MVM.
We have shown that the synthetic Emphaze device’s excellent viral clearances for both MVM and X-MuLV (3.7–5.5 LRV) are driven by electrostatic adsorption. These results are in accordance with previously reported results obtained for another MAb, suggesting robust viral clearance (8). They also support the mechanistic operation mode of anion-exchange modes, which operate almost exclusively by charge-based separation considered to be robust in terms of viral clearance (16). Our results also could be verified by HCP removal results, which confirmed an electrostatic interaction as the only retention mechanism for the fully synthetic device. Furthermore, results of a previous virus-spiking study of a conventional Q-functionalized membrane adsorber showed no significant MVM removal even with a 4.5× lower MAb load under the same buffer-system conditions (data not published).
We hypothesize that the Emphaze AEX Hybrid Purifier device can withstand the concentration of polyvalent ions in a buffer system through its high ion-exchange capacity, offering a significant advantage over conventional Q-functionalized filter media. The X-MuLV clearance of 5 LRV by the nonwoven component of the Emphaze filter is higher than the published average X-MuLV clearance of 2–4 LRV by AEX resins and adsorbers (16). Key process economic advantages include higher flow rates, reduced process time, disposability, and lower buffer consumption.
References
1 Miesegaes G, et al. Analysis of Viral Clearance Unit Operations for Monoclonal Antibodies. Biotechnol. Bioeng. 106(2) 2010: 238–246; doi:10.1002/bit.22662.
2 ICH Q5A. Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human of Animal Origin. US Fed. Reg. 63(185) 1998: 51074.
3 Brown M, et al. A Step-Wise Approach to Define Binding Mechanisms of Surrogate Viral Particles to Multi-Modal Anion Exchange Resin in a Single Solute System. Biotechnol. Bioeng. 114(7) 2017: 1487–1494; doi:10.1002/bit.26251.
4 Shukla A, et al. Viral Clearance for Biopharmaceutical Downstream Processes. Pharm. Bioprocess. 3(2) 2015: 127–138.
5 Shukla A, et al. Downstream Processing of Monoclonal Antibodies: Application of Platform Approaches. J. Chromatogr. B 848(1) 2007: 28–39; doi:10.1016/j.jchromb.2006.09.026.
6 Yinges Y, et al. Exploitation of the Adsorptive Properties of Depth Filters for Host Cell Protein Removal During Monoclonal Antibody Purification. Biotechnol. Prog. 22(1) 2006: 288–296; doi:10.1021/bp050274w.
7 Singh N, et al. Development of Adsorptive Hybrid Filters to Enable Two-Step Purification of Biologics. MAbs 9(2) 2017: 350–364; doi:10.1080/19420862.2016.1267091.
8 Metzger M, et al. Evaluating Adsorptive Filtration As a Unit Operation for Virus Removal. BioProcess Int. 13(2) 2015: 36–44.
9 Venkiteshwaran A, et al. Mechanistic Evaluation of Virus Clearance By Depth Filtration. Biotechnol. Prog. 31(2) 2015: 431–437; doi:10.1002/btpr.2061.
10 Zhou JX, et al. Viral Clearance Using Disposable Systems in Monoclonal Antibody Commercial Downstream Processing. Biotechnol. Bioeng. 100(3) 2008: 488–496; doi:10.1002/bit.21781.
11 Wang M. Zeta + VR Filters for Viral Reduction. BioProcess Int. 9(7) 2011: 62.
12 Zhou JX, et al. Orthogonal Virus Clearance Applications in Monoclonal Antibody Production. Process Scale Purification of Antibodies. Gottschalk U, Ed. John Wiley and Sons: Hoboken, NJ, 2009: 169–186.
13 Trilisky E, et al. Flow-Dependent Entrapment of Large Bioparticles in Porous Process Media. Biotechnol. Bioeng. 104(1) 2009: 127–133; doi:10.1002/bit.22370.
14 Singh N, et al. Clarification Technologies for Monoclonal Antibody Manufacturing Processes: Current State and Future Perspectives. Biotechnol. Bioeng. 113(4) 2016: 698–716; doi:10.1002/bit.25810.
15 Michen B, et al. Virus Removal in Ceramic Depth Filters Based on Diatomaceous Earth. Environ. Sci. Technol. 46(2) 2012: 1170–1177; doi:10.1021/es2030565.
16 Miesegaes G, et al. Viral Clearance By Flow-Through Mode Ion-Exchange Columns and Membrane Adsorbers. Biotechnol. Prog. 30(1) 2014: 124–131; doi:10.1002/btpr.1832.
Corresponding author Anja Trapp ([email protected]) is a scientist in bioprocessing technology and innovation, Laura Igl is a technical assistant, Sabine Faust is a process engineer, Alexander Faude is group leader, Roland Wagner is senior advisor and head of production, and Stefan Schmidt is chief scientific officer at Rentschler Biopharma SE in Laupheim, Germany. Corresponding author Dr. Sophie Muczenski is a specialist application engineer, and Nicole Mang is a sales account executive in the separation and purification sciences division at 3M Deutschland GmbH, Carl-Schurz-Straße 1, 41453 Neuss, Germany; 49-2131-145133; [email protected]. Zeta Plus and Emphaze are registered trademarks of 3M. AKTAexplorer and Unicorn are registered trademarks of GE Healthcare.
You May Also Like





