Creating Novel Cell Lines By Genome Editing: Simplifying Cell-Based Assays and Improving Production of BiomoleculesCreating Novel Cell Lines By Genome Editing: Simplifying Cell-Based Assays and Improving Production of Biomolecules
September 26, 2019
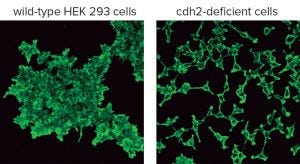
Figure 1: HEK293 wild-type and cdh2-deficient cells were stained with fluorescein-conjugated phalloidin (2).
Cultured cell lines have a diverse range of applications. They are used broadly by cell biologists, clinicians, tissue engineers, biotechnology scientists, and bioengineers. The most important uses of cell culture are in the cell-based assays and production of biologically active recombinant proteins. In recent years, genome editing has been used widely to study the structure, function, and localization of endogenous proteins in cultured cells. However, applying the same genome editing techniques to cell lines also could improve the propagation of cells and help scientists devise novel assays for discovering bioactive molecules. Below, using a cadherin adhesion molecule as an example, I explain a simple approach to genome editing for generating novel cell lines to achieve those goals.
Case Study Cadherins are cell-surface transmembrane proteins that mediate cell-to-cell adhesion in animals. In 1987, Masatoshi Takeichi and colleagues at Kyoto University introduced cDNA of E-cadherin (CDH1) into fibroblastic cells lacking intrinsic cadherin (1). The transfected fibroblasts stuck together and aggregated. That simple experiment proved that cadherins are molecules required for intercellular adhesion.
Cadherins are cell-surface transmembrane proteins that mediate cell-to-cell adhesion in animals. In 1987, Masatoshi Takeichi and colleagues at Kyoto University introduced cDNA of E-cadherin (CDH1) into fibroblastic cells lacking intrinsic cadherin (1). The transfected fibroblasts stuck together and aggregated. That simple experiment proved that cadherins are molecules required for intercellular adhesion.
Human embryonic kidney (HEK293) cells are one of the most widely used cell lines in biomedical research as well as industrial production of therapeutic molecules and viral vectors. The cell line is easy to transfect efficiently and grows well in culture. HEK293 cells have strong intrinsic cell–cell adhesion mediated by cadherin-2 (CDH2, N-cadherin). By applying CRISPR/Cas9 technology (see box, right), we recently deleted the cdh2 gene from HEK293 cells. The resulting cdh2-deficient HEK293 cells do not aggregate (Figure 1) (2). By overexpressing the wild-type cdh2 gene, then CDH2-mediated adhesion can be rescued.
Here’s the advantage of removing cell–cell adhesion from cultured cells: Passage and maintenance in culture of cdh2-deficient HEK293 cells become effortless without the need for trypsin to dissociate them. It could be possible to culture them under unique conditions (e.g., adherent but higher density culture, or suspension culture) because they do not aggregate.
Novel HEK293-cell–based assays could be created for discovery and study of protein–protein interactions. This cell line is suited particularly well for such experiments because it is highly transfectable with exogenous DNA. For example, the specificity of cell-adhesion molecules can be tested in cdh2-deficient cells that lack cell–cell adhesion activity. The cell line can be used as a reporter system for monitoring protein–protein interactions on cell surfaces that otherwise would be hampered by CDH2-mediated adhesion. We showed that the structure and function of several synaptic-adhesion molecules can be studied by transfecting this cell line (2, 3).
Getting Rid of Annoying Genes
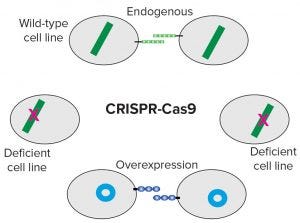
Figure 2: Elimination of endogenous genes that potentially hamper overexpressed genes reveals the activity of overexpressed genes.
Figure 2 shows how the idea can be extended. Overexpression once was considered to be predominantly a method for increasing the productivity of cultured cell lines. By introducing plasmids or riboproteins to generate CRISPR/Cas9 complexes into such lines, their genomes can be edited at any desired position (4, 5). This genome-editing technique has been used widely to study the structure, function, and localization of endogenous proteins both in vivo and in cultured cells. By applying genome editing to cell lines, we can design them simply by eliminating troublesome or annoying genes.
 For example, the genes that mask particular protein–protein interactions can be eliminated from a given cell line. This enables the creation of novel cell-based assay systems for studying exogenous proteins by overexpression using the resulting mutant cell line. If a cell line expresses endogenously either inhibitors or activators of a protein of interest, you can eliminate them by using this straightforward technique. It might sound simple, but it opens up a whole new avenue for the future use of cell lines.
For example, the genes that mask particular protein–protein interactions can be eliminated from a given cell line. This enables the creation of novel cell-based assay systems for studying exogenous proteins by overexpression using the resulting mutant cell line. If a cell line expresses endogenously either inhibitors or activators of a protein of interest, you can eliminate them by using this straightforward technique. It might sound simple, but it opens up a whole new avenue for the future use of cell lines.
References
1 Nagafuchi A, et al. Transformation of Cell Adhesion Properties By Exogenously Introduced E-Cadherin cDNA. Nature 329(6137) 1987: 341–343; doi:10.1038/329341a0.
2 Yamagata M, Duan X, Sanes JR. Cadherins Interact with Synaptic Organizers to Promote Synaptic Differentiation. Front. Molec. Neurosci. 11, 30 April 2018: 142; doi:10.3389/fnmol.2018.00142.
3 Goodman KM, et al. Molecular Basis of Sidekick-Mediated Cell–Cell Adhesion and Specificity. eLife 5,19 September 2016; doi:10.7554/eLife.19058.
4 Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157(6) 2014: 1262–1278; doi:10.1016/j.cell.2014.05.010.
5 Knott GJ, Doudna JA. CRISPR-Cas Guides the Future of Genetic Engineering. Science 361(6405) 2018: 866–869; doi:10.1126/science.aat5011. c
Masahito Yamagata, PhD, is a Center for Brain Science researcher in the Department of Molecular and Cellular Biology at Harvard University, 52 Oxford Street, Cambridge, MA 02138; 1-617-496-6868; [email protected].
You May Also Like






