Application of Omics Technologies: Creating Next-Generation CHO Expression PlatformsApplication of Omics Technologies: Creating Next-Generation CHO Expression Platforms
Biomanufacturing platforms based on Chinese hamster ovary (CHO) cells have transformed the industrial production of biologics such as monoclonal antibodies (mAbs) over the past few decades. Despite advances in alternative expression systems (e.g., microbial, insect, other mammalian cells, and cell-free synthesis), the dominance of the CHO platform in the biopharmaceutical industry continues. As the therapeutic protein landscape continues to evolve, the fundamental requirements of biomanufacturing processes remain consistent: An economically viable CHO-based bioprocess requires stably high titers of expressed recombinant protein with controlled product quality (PQ).
Historically, advancements in CHO cell-line productivity have been driven predominantly by process optimization rather than direct genetic manipulation of cells themselves. Early efforts in biopharmaceutical manufacturing focused on refining production parameters such as culture conditions, feeding strategies, bioreactor design, and purification techniques to boost yields and reduce costs. Those process-oriented improvements enabled manufacturers such as Lonza to scale up commercial production and enhance the consistency of biologics. And although such strategies delivered significant gains, they simultaneously highlighted a growing need to explore fundamental vector-based and cell-line–specific modifications to sustain productivity improvements and meet the evolving requirements of modern biotherapeutics.
Consider the evolution of CHO fed-batch titers for mAbs, which realized substantial increases in titers from the early 1990s through today (Figure 1). The rate of titer increase slowed in recent years as expression platforms matured and priorities shifted somewhat toward speed, agility, and PQ. For the future, we expect diverse pressures on biomanufacturing platforms that reflect both existing challenges and new ones linked to societal shifts. Advances in personalized medicine and the development of intricate biologics require cell lines that can deliver consistently high yields with reduced production times and lower costs. Additionally, environmental sustainability concerns and regulatory requirements are driving the need for more efficient use of resources, reduced waste, and improved product quality.
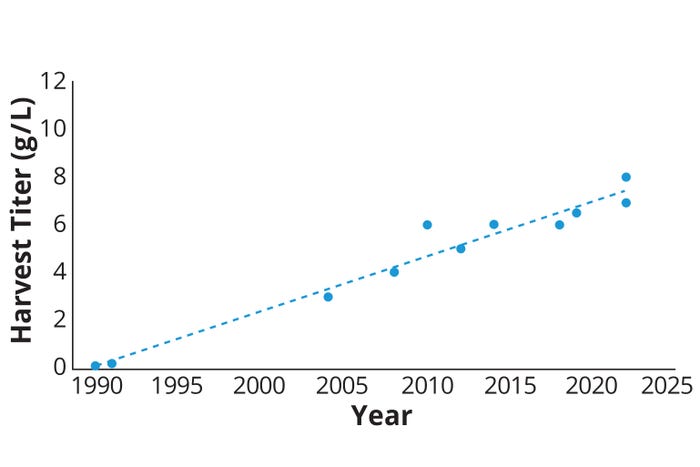
Figure 1: Trends in production titers from glutamine-synthetase– knockout Chinese hamster ovary (GS-CHO) platforms (based on in-house and published Lonza data) from the 1990s to the present.
Clearly, the next generation of CHO-based bioproduction platforms will have to contend with a broadly diverse set of challenges. So it’s worth considering what technologies the industry has at its disposal to meet those demands. Below, we consider how “omics” technologies can be used to create improved expression platforms. Then we suggest where the application of predictive tools linked to omics data sets could help to create truly transformative expression solutions in the future.
Building a Next-Generation Expression Platform
Figure 2 shows some key ingredients required to create a next-generation CHO-based bioprocess. Final product titer and overall yield are key elements of every bioprocess — along with PQ — but are not the only requirements. Although typical CHO productivity rates can vary from ~20–70 pg/cell/day, professional secretory cells (e.g., human B cells), purportedly can achieve rates up to ~180 pg/cell/day (1). Based on the assumption that CHO cells contain all the necessary genetic information and physiological apparatus for reaching such lofty goals, we believe that the productivity ceiling in CHO has yet to be reached. The obvious question becomes how to reach such productivity rates consistently for a wide range of recombinant protein product types.
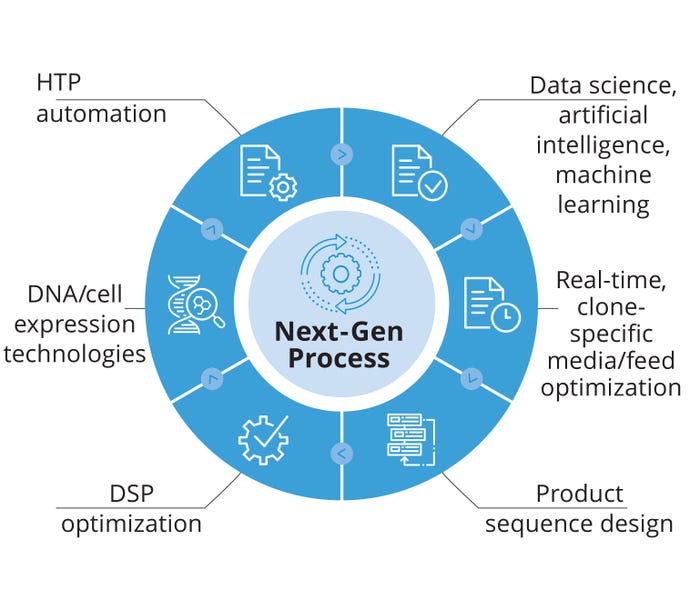
Figure 2: Key ingredients required to construct a next generation CHO-based bioproduction platform; HTP = high throughput, DSP = downstream processing.
Focusing specifically on DNA and cell-expression technologies (Figure 2), we believe that an improved understanding of the underlying CHO cell “factory” is of the utmost importance for reaching the productivity targets alluded to above. Such understanding begins with characterizing clones at a genetic level, then moves on through multiple phases of cellular regulation that result in the production of proteins. Using a systems approach, we seek to achieve a holistic understanding of CHO performance within an industrial environment. That in turn can serve as the basis for targeted process-engineering approaches.
Brief History of CHO Genomics
To build a holistic, omics-led understanding of the CHO cell factory, we first need a high-quality, complete reference genome assembly. Despite the huge health and economic impact of CHO-based expression tools, CHO cell lines came relatively late to the genomics party, mostly due to a lack of industry–academic coordination and the fragmented nature of the user base. Numerous derivatives of the original Puck isolates now are in widespread circulation and use (2). These challenges no doubt were exacerbated by the highly plastic and malleable nature of the cell lines themselves — a useful attribute for expressing diverse protein formats in industrial biomanufacturing processes based on screening, but less beneficial when it comes to establishing a baseline reference genome.
The first draft genome sequence of the CHO-K1 cell line, the progenitor of many industrial platform cell lines, was published in 2011 (3). The associated full hamster genome and other CHO lines were published soon afterward (4, 5). After iterative updates as new sequencing platforms and bioinformatics pipelines arrived, the current standard assembly for the Chinese hamster is the chromosome-scale “PICRH” assembly that was built using improved Hi-C enabled scaffolding, among other advances, and published in 2020 (6).
The original 2.45-Gb assembly presented in 2011 delivered a number of novel insights, including into key cellular performance attributes such as the genetic basis of CHO’s renowned human-like glycosylation and the fact that the genome boasts nearly 38% transposable elements (3). Explosive growth has followed in the use of transposases in CHO-based manufacturing (7).
With the Lewis et al. and Brinkrolf et al. publications (4, 5), it became possible to uncover some of the genetic diversity that exists not only between the Chinese hamster and derivative CHO cell lines, but also between lineages such as CHO-S, CHO DG44, and different CHO-K1 descendants. Of particular note herein was the identification of single-nucleotide polymorphisms (SNPs) specific to recombinant-antibody–derivative CHO cell lines. SNPs helped to elucidate data at nucleotide-level resolution to support oft-repeated statements about the rapid plasticity of the CHO genome after transfection. Correlating such changes (and larger-scale rearrangements) with clone performance should help scientists identify those specific to phenotypes that are desirable for production platforms.
Across the bioprocessing industry, several proprietary CHO cell lines have been sequenced and annotated using the latest tools (8, 9). Those developments reflect the need for platform-specific data sets. Although such data typically are not available for widespread public consultation, they nonetheless add to the cumulative CHO genomics knowledge across the bioprocessing community and help to expand the availability of reference standards.
CHO Genome Analysis at Lonza: Differences among alternative CHO hosts and between each host and the reference hamster PICRH genome necessitate platform-specific assemblies to support activities such as cell engineering. Indeed, harnessing the genome blueprint is an essential first step for detailed molecular characterization of clones and for precise cell-engineering approaches that go beyond single-gene edits.
Our company has developed industry-leading omics data sets aligned to the unique provenance of the platform GS Xceed CHOK1SV GS-KO cell line (Figure 3). Using the latest technologies, notably long-read sequencing using nanopore (a third-generation sequencing platform), we have created a de novo assembly of the genome with quality metrics such as genome completeness using the benchmarking universal single-copy orthologs metric (BUSCO, https://busco.ezlab.org) equivalent to relevant genomes published in the public domain (Figure 4).
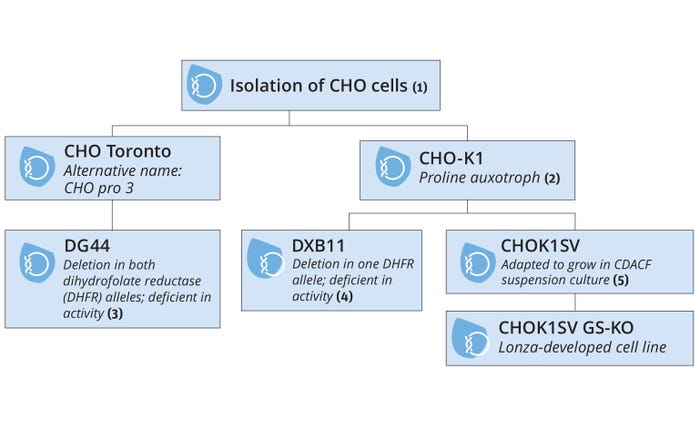
Figure 3: History and provenance of the Lonza CHOK1SV GS-KO host-cell line; CDACF = chemically defined, animal-component free.
1 Puck TT, Cieciura SJ, Robinson A. Genetics of Somatic Mammalian Cells. III. Long-Term Cultivation of Euploid Cells from Human and Animal Subjects. J. Exp. Med. 108, 1958: 945–956; https://doi.org/10.1084/em.108.6.945.
2 Kao FT, Puck TT. Genetics of Somatic Mammalian Cells, VII. Induction and Isolation of Nutritional Mutants in Chinese Hamster Cells. Proc. Nat. Acad. Sci. USA 60(4) 1968: 1275–1281; https://doi.org/10.1073/pnas.60.4.1275.
3 Urlaub G, et al. Deletion of the Diploid Dihydrofolate Reductase Locus from Cultured Mammalian Cells. Cell 33(2) 1983: 405–412.; https://doi.org/10.1016/0092-8674(83)90422-1.
4 Urlaub G, Chassin LA. Isolation of Chinese Hamster Cell Mutants Deficient in Dihydrofolate Reductase Activity. PNAS 77(7) 1980: 4216–4220; https://doi.org/10.1073/pnas.77.7.4216.
5 Rendall M. et al. Transfection to Manufacturing: Reducing Timelines for High Yielding GS-CHO Processes. Gòdia F, Fussenegger M, Eds. Animal Cell Technology Meets Genomics: ESACT Proceedings 2. Springer: Dordrecht, Germany, 2005; https://doi.org/10.1007/1-4020-3103-3_144.
The nucleotide “nuts and bolts” are critical, of course, but accurate and high-quality annotations are key to making a reference genome useful for meaningful work. A common method involves using reference-gene lists to annotate genomes with automated bioinformatics tools followed by bespoke manual curation to account for divergent species and genomes. This has been particularly challenging with earlier, more fragmented CHO assemblies that can include errors carried over from draft genomes — and cross-species comparisons come with significant gaps. At Lonza, we have put substantial effort into correcting the outputs from automated gene-prediction models to achieve a well-annotated genome that can support precise, nucleotide-level gene-editing work. To further improve the quality of our builds, we are future-proofing our genomes by building high-throughput, automated pipelines based on cloud computing for easy integration of additional information as it becomes available.
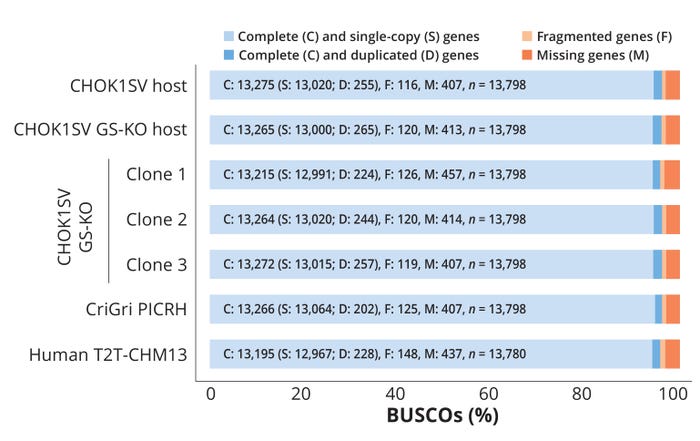
Figure 4: Comparing the completeness of Lonza’s in-house Chinese hamster ovary (CHO) genome assemblies with that of public reference genomes using benchmarking universal single-copy orthologs (BUSCO) scores; n = number of genes.
Holistic Omics Analysis of CHO Cell Lines: Accurate and representative genome maps of biomanufacturing host-cell lines are needed to enable targeted cell engineering efforts for improved performance attributes (among other goals). However, meaningfully linking genome sequences with output CHO cell phenotypes requires a deep understanding of the relationship between the cellular nucleotide “hardware” and the expression “firmware/software.” Toward such ends, we can apply epigenomics, transcriptomics, proteomics, metabolomics, and so on — all areas of study that have exploded in interest within the past decade or so.
Our company has invested significantly in building out multilevel omics knowledge of our host-cell lines, with a particular interest in exploring the three-dimensional (3D) genome space (9). Working with academic and industrial partners, Lonza pioneered the study of CHO 3D genome structures — using techniques such as Promoter Capture Hi-C (10) combined with Hi-C, ATAC-Seq (11), and traditional ChIP-Seq data related to histone modifications (12) — to interrogate genome architecture within host and recombinant clonal cell lines used to support research and development (Figure 5). With such data sets, we can delve deeply into the mechanisms underpinning divergent CHO cell phenotypes and begin to explore how broadly similar CHO genomes can generate such diverse phenotypes.
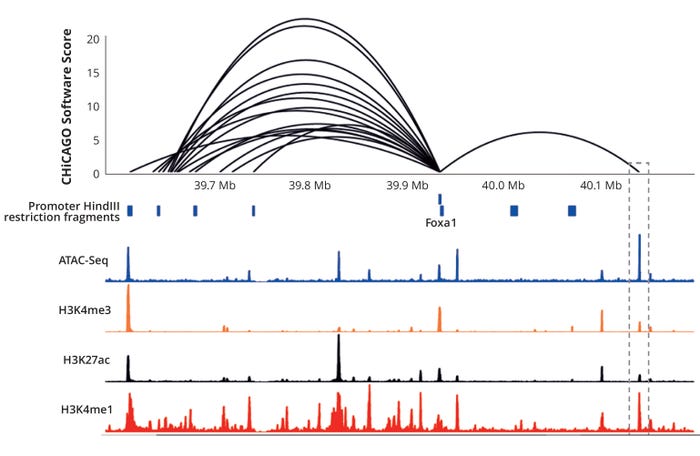
Figure 5: Location of the Foxa1 gene relative to cis-promoter interactions in a GS-CHO cell line is revealed by promoter capture Hi-C (PCHi-C), open chromatin (ATAC-Seq), and common epigenetic marks indicative of active chromatin (9); H3K4me3 = trimethylation of histone H3 at lysine 4; H3K27ac = acetylation of lysine 27 on histone H3; H3K4me1 = monomethylation of lysine 4 of histone H3.
To relate such genetic and epigenetic maps to cellular function, however, further omics data sets are required — RNASeq, for example. Lonza applied lessons learned from the above-referenced studies to create comprehensive, multilevel omics maps of the proprietary GS Xceed CHOK1SV GS-KO host-cell line. When combined with in-house bespoke software tools — based on the Integrative Genomics Viewer (IGV) desktop application — those maps enable visualization of every gene in the overall sequence along with the associated 3D interactome, enhancer-activity maps (made with the self-transcribing active regulatory region sequencing (STARR-Seq) method), and RNA-sequencing expression readouts (Figure 6). Using the resulting data sets, we could begin to understand how individual genes achieve their characteristic expression levels, and are influenced by nearby (and not so nearby) sequence elements with functions that were hitherto unknown to us.
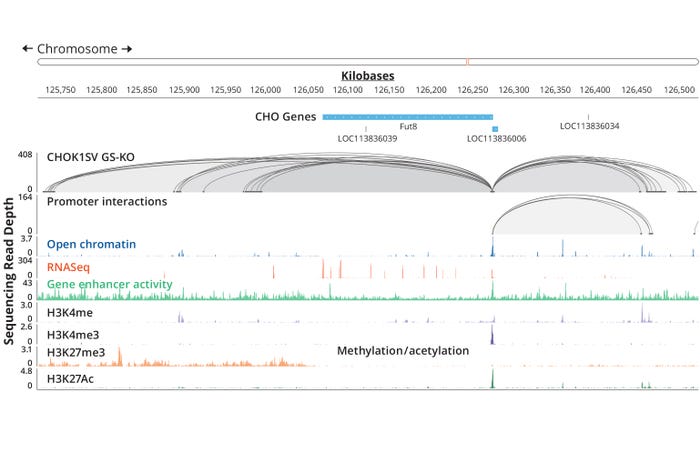
Figure 6: Visualizing the Fut8 gene within the GS Xceed CHOK1SV GS-KO genome assembly and associated data sets using an in-house platform; promoter interactions were revealed by Promoter Capture Hi-C (PCHi-C) using HindIII and DpnI; open chromatin was revealed by ATAC-Seq; gene enhancer activity was revealed by STARR-Seq. Methylation (me) and acetylation (ac) of histone H3 on lysines 4 and 27 are indicated.
For increasingly in-depth insight, we use single-cell omics analyses to provide high-resolution information about individual cell behaviors and changes that bulk cell-population studies inevitably amalgamate or obscure (13). Those data sets should help us to optimize manufacturing processes further. In industrial recombinant protein expression by CHO cells, for example, such analyses could help researchers understand and mitigate production instability by identifying genetic and epigenetic factors contributing to variability, leading to the selection of more robust clones (14). The resulting information enables examination of endoplasmic reticulum (ER) stress (15) and mitochondrial DNA heterogeneity across cell populations (16), both being important factors for the production of complex proteins.
Ultimately, single-cell technologies will enhance analytical throughput with multiplexed experiments that enable multiple clonal cell lines to be cocultured in the same environment. That approach not only increases throughput, but also provides a robust experimental setup for discovery and facilitates the simultaneous study of different cell lines under identical conditions. We believe that single-cell markers of productivity and stability ultimately will play a crucial role in optimizing bioprocessing techniques. They will improve the precision of selection for high-performing clones and advance the development of robust and efficient therapeutic protein-production systems.
Using Multilevel Omics Data for Improved Expression Outcomes: Building comprehensive multilayered omics maps as described above can deepen our understanding of the CHO cell protein-production machinery and provide essential elements for prescriptive analysis. This goes beyond so-called “academic” interest in interrogating the biological functions of the biopharmaceutical industry’s most critical “workhorse.” We also ask how to turn such insights into meaningful outcomes that enable the industry to meet growing patient demand for biotherapeutics while reducing cost of goods (CoG) for ever more diverse protein formats.
Broadly speaking, we can move forward with three steps:
• identification of predictive omics markers to accelerate the detection and analysis of fundamental cellular attributes linked to metrics such as expression titer, genetic stability, and product quality
• use of predictive omics markers to increase testing throughput and maximize successful development of improved expression technologies (cells, expression vectors, media, and so on)
• use of predictive omics markers to help us understand CHO cells’ protein-production machinery and design prescriptive tools to improve overall bioprocess operations.
The development of omics tools enables platform improvements, most obviously by accelerating our detection of productivity attributes and targets for cell engineering. Improved genome assemblies can facilitate the design of direct gene-editing approaches (e.g., guide ribonucleic acid (gRNA) design) and also could enable subtle interventions such as editing elements of the interactome to influence genome readout (and therefore phenotype) indirectly. The ability to identify genome sequences with “super-enhancer” activity (17) could help to improve expression vectors, which have remained fairly consistent in design over the past couple of decades.
Additionally, deep-learning approaches aligned to the omics data sets described above could be used to design bespoke enhancers that help us to achieve specific expression outcomes, such as temporal control. Application of epigenome editing could improve control over transcriptional outputs within CHO cell factories (18), perhaps to unleash further the somewhat constrained secretory capacity of cell lines that trace their origins backed to nonsecretory progenitors.
In parallel with the above, a possibly more powerful expression of the potential inherent to such collective data sets would be their use in building predictive tools for platform improvement. Each individual cell at the point of clonal isolation should contain within itself — even at the primary stage of cell-line construction (CLC) — the necessary biological information required to predict that cell’s suitability as a lead clone to support a manufacturing campaign. Biological systems are inherently variable and subject to change. But equipped with enough datapoints and the application of sufficiently powerful machine-learning (ML)/artificial-intelligence (AI) tools, we theoretically could select lead clones much earlier in the CLC workflow without full end-to-end empirical screening (Figure 7).
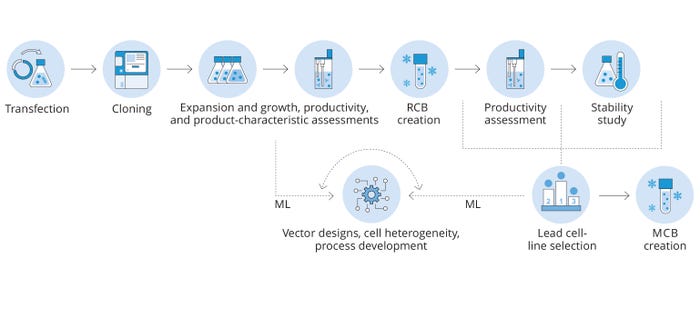
Figure 7: Based on a sufficiently large set of multilevel omics data, we anticipate that application of machine learning (ML) ultimately will lead to greatly enhanced lead-clone prediction, potentially at the earliest single-cell stage; MCB = master cell bank, RCB = research cell bank.
Current work at Lonza is leveraging our expertise in omics and vector engineering to identify early on the optimal combinations of factors to yield clones exhibiting desired phenotypes across a production process. In addition to focusing on the omics aspects, we also are developing and refining the necessary tools and algorithms to support that work. They include advanced ML frameworks that can handle the large number of features present in multiomics data sets. By integrating sophisticated computational approaches with our biological research, we intend to decrease as much as possible the biologically derived randomness that has governed production processes in the past, essentially evolving what we can call “precision bioprocessing.”
Identification of a comprehensive molecular “fingerprint” for CHO bioprocessing may be several years away and influenced to some degree by product-specific effects. However, our efforts toward that “blue sky” goal undoubtedly will yield significant short- to medium-term benefits. As we focus on fundamental cellular attributes that correlate broadly with improved expression outcomes, we are poised to make substantial progress and drive immediate advancements in our bioprocessing capabilities.
A Journey of Discovery — and Development
Development of CHO omics has come a long way in recent years and in very broad terms is beginning to approach parity (in quality and depth) with data sets from other systems such as mouse and human. The proprietary nature of CHO expression platforms inevitably leads to an “individualistic” array of data sets that hampers community-wide use for cell engineering and other generic system-optimization efforts. That could be considered limiting in the broad sense, but it has led companies such as Lonza to develop highly specific, detailed, and comprehensive sets that can be used to enhance CHO platforms that already have reached a high level of performance optimization and maturity.
We anticipate that the omics tools described herein will enable a step-change platform upgrade for the biopharmaceutical industry. The ultimate expression will come in the development of prescriptive tools that deliver on-demand, real-time predictions of DNA–host–process combinations at the single-cell stage, thereby eliminating much of the current CLC workflow. As we work toward that goal, interim outputs focused on engineered CHO host cell lines and enhanced expression-vector designs will help to drive down CoG for a wide array of product types.
While focusing on the ultimate end goal, we mustn’t forget the pleasure of the journey. Rather than seeking to fly over the waypoints, curiosity-driven, multidisciplinary scientists look forward to the myriad discoveries coming over the next few years and welcome the unique insights that omics and related tools can bring.
Acknowledgment
We are grateful to Dr. Daniel Fabian for critical review of this manuscript.
References
1 Henn A, et al. Modulation of Single-Cell IgG Secretion Frequency and Rates in Human Memory B Cells By CpG DNA, CD40L, IL-21, and Cell Division. J. Immunol. 183, 2009: 3177–3187; https://doi.org/10.4049/jimmunol.0804233.
2 Puck TT, Cieciura SJ, Robinson A. Genetics of Somatic Mammalian Cells: III. Long-Term Cultivation of Euploid Cells from Human and Animal Subjects. J. Exp. Med. 108(6) 1958: 945–956; https://doi.org/10.1084/jem.108.6.945.
3 Xu X, et al. The Genomic Sequence of the Chinese Hamster Ovary (CHO) K1 Cell Line. Nat. Biotechnol. 29(8) 2011: 735–741; https://doi.org/10.1038/nbt.1932.
4 Lewis NE, et al. Genomic Landscapes of Chinese Hamster Ovary Cell Lines As Revealed By the Cricetulus griseus Draft Genome. Nat. Biotechnol. 31(8) 2013: 759–765; https://doi.org/10.1038/nbt.2624.
5 Brinkrolf K, et al. Chinese Hamster Genome Sequenced from Sorted Chromosomes. Nat. Biotechnol. 31(8) 2013: 694–695; https://doi.org/10.1038/nbt.2645.
6 Hilliard W, MacDonald ML, Lee KH. Chromosome-Scale Scaffolds for the Chinese Hamster Reference Genome Assembly To Facilitate the Study of the CHO Epigenome. Biotechnol. Bioeng. 117(8) 2020: 2331–2339; https://doi.org/10.1002/bit.27432.
7 Rajendra Y, Peery RB, Barnard GC. Generation of Stable Chinese Hamster Ovary Pools Yielding Antibody Titers of Up to 7.6 g/L Using the piggyBac Transposon System. Biotechnol. Prog. 32(5) 2016: 1301–1307; https://doi.org/10.1002/btpr.2307.
8 Kretzmer C, et al. De Novo Assembly and Annotation of the CHOZN® GS−/− Genome Supports High-Throughput Genome-Scale Screening. Biotechnol. Bioeng. 119, 2022: 3632–3646; https://doi.org/10.1002/bit.28226.
9 Bevan S, et al. High-Resolution Three-Dimensional Chromatin Profiling of the Chinese Hamster Ovary Cell Genome. Biotechnol. Bioeng. 118(2) 2021: 784–796; https://doi.org/10.1002/bit.27607.
10 Schoenfelder S, et al. The Pluripotent Regulatory Circuitry Connecting Promoters to Their Long-Range Interacting Elements. Genome Res. 25(4) 2015: 582–597; https://doi.org/10.1101/gr.185272.114.
11 Buenrostro JD, et al. Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 10, 2013: 1213–1218; https://doi.org/10.1038/nmeth.2688.
12 Feichtinger J, et al. Comprehensive Genome and Epigenome Characterization of CHO Cells in Response to Evolutionary Pressures and Over Time. Biotechnol. Bioeng. 113(10) 2016: 2241–2253; https://doi.org/10.1002/bit.25990.
13 Pilbrough W, Munro TP, Gray P. Intraclonal Protein Expression Heterogeneity in Recombinant CHO Cells. PLoS One 4(12) 2009: e8432; https://doi.org/10.1371/journal.pone.0008432.
14 Tzani I, et al. Tracing Production Instability in a Clonally Derived CHO Cell Line Using Single-Cell Transcriptomics. Biotechnol. Bioeng. 118(5) 2021: 2016–2030; https://doi.org/10.1002/bit.27715.
15 Tzani I, et al. Understanding the Transcriptional Response to ER Stress in Chinese Hamster Ovary Cells Using Multiplexed Single Cell RNA-seq. bioRxiv 31 March 2022; https://doi.org/10.1101/2022.03.31.486542.
16 Foley A, et al. A Complete Workflow for Single Cell mtDNAseq in CHO Cells, from Cell Culture to Bioinformatic Analysis. Front. Bioeng. Biotechnol. 12, 2024: 1304951; https://doi.org/10.3389/fbioe.2024.1304951.
17 Hnisz D, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell 155(4) 2013: 934–947; https://doi.org/10.1016/j.cell.2013.09.053.
18 Policarpi C, et al. Systematic Epigenome Editing Captures the Context-Dependent Instructive Function of Chromatin Modifications. Nat. Genet. 56(6) 2024: 1168–1180; https://doi.org/10.1038/s41588-024-01706-w.
Corresponding author Peter M. O’Callaghan is senior director and head of expression system sciences and licensing, and Alessandro Di Cara is head of bioinformatics and data science R&D, both at Lonza Biologics, 234 Bath Road, Slough SL1 4DX, UK; 44-1753-777000; [email protected]; https://www.lonza.com.
GS Xceed and CHOK1SV GS-KO are registered trademarks of Lonza Group AG.
You May Also Like







