Pathogen Safety Digital Platform for Biopharmaceuticals: The Journey from Ground to CloudPathogen Safety Digital Platform for Biopharmaceuticals: The Journey from Ground to Cloud
Digital transformation is at the heart of many biopharmaceutical companies’ strategies for ongoing success. However, the definition of digital transformation and what it consists of differ within the bioprocess industry (and might even vary within a single company), specifically as it applies to the overall value chain from R&D to clinical trials, manufacturing, supply chain, and eventually commercial operations. To provide a perspective on what digital transformation could look like in bioprocessing, we present a case study about an exploratory innovation project that showcases the application of digital twins in bioprocess development and other industries (e.g., software development).
Digital twins are at the core of many new approaches in digitizing nontechnical industries. Negri et al. (1) state that a digital twin is “meant as the virtual and computerized counterpart of a physical system that can be used to simulate it for various purposes, exploiting a real-time synchronization of the sensed data coming from the field.” The core concept is that a model exists to represent the behavior of a system under consideration. The type of model used as a basis for a digital twin can be first principle, data driven, or a hybrid of both. In the context of biopharmaceutical process development, digital twins can help streamline and accelerate experiments associated with product commercialization. From an industry perspective, the possibility of reducing the number of time- and resource-intensive wet-laboratory experiments serves as a strong business driver.
Software platforms play important roles in digital-twin implementation. In the past couple of decades, many open-source and commercial cloud-based software platforms have emerged. These are alternatives to traditional desktop-based software for statistical analysis and mathematical modeling. Because of the cloud-based nature of these software, computational power is no longer a limitation. Open-source software enables users to make modifications as needed. Overall, the new generation of software platforms provides improved flexibility and user experience.
Historically, biopharmaceutical companies were driven by static processes and procedures. That affected the way software and other applications were developed and implemented — with a lot of upfront planning and waterfall-like execution of a project and delivery of specific user requirements. However, that approach does not fit into the nature of exploratory projects, so the project team for the case study below decided to use a more flexible approach. Although they did not completely embrace every aspect of agile development, they followed the basic principles. The general guidance was to work in short cycles (sprints) that produced working solutions and underwent regular end-user feedback.
Based on guidelines from the US Food and Drug Administration (FDA) and the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) (2–4), biopharmaceutical companies are expected to enhance their scientific understanding of their products and processes, ensure product quality and process performance by design, and gain knowledge of life-cycle management. Some business drivers include shortening time to market, reducing cost of goods (CoG), and increasing manufacturing capacity. To meet such compliance and business objectives, biomanufacturers need to complete activities related to scientific, engineering, operational, and business requirements that span product and process life cycles. Examples of such activities include drug-candidate manufacturability assessment, process characterization, product specification setting, scale-up, technology transfer, continued process verification, and postlaunch improvements.
Digital twins can leverage available R&D and manufacturing data to help subject-matter experts (SMEs) make well-informed, data-driven organizational decisions. Digital twins also can help companies meet their compliance and business objectives by enhancing scientific understanding and control of biomanufacturing processes and streamlining the process of obtaining knowledge from underlying data. For example, process characterization and optimization during product development can be streamlined with the use of mathematical models. Those can be leveraged to understand relationships among process parameters and quality attributes and to determine optimal process conditions, all with fewer wet-laboratory experiments than typically would be required.
To realize fully the benefits of digital twins, the biopharmaceutical industry needs to embrace best practices, data science methods, digital technologies, and learnings from other industries (e.g., the software industry). Some steps that biomanufacturers can take to advance toward digitalization include
• adopting technologies such as cloud platforms
• establishing cross-functional teams of SMEs, data scientists, cloud experts, and project managers
• applying methodologies typically found in the software industry such as agile product development.
Case Study
To provide our perspective on the implementation of digital twins in process development, we selected the pathogen clearance case study below. This selection was based on availability of internally generated experimental data and publicly available data through a pharmaceutical-company consortium. The intent is to share learnings and experience that could be helpful for biotechnology companies for the advancement of digital transformation.
Case Study for Biological Process Development: Most recombinant therapeutic proteins are manufactured by large-scale culture of animal- or human-derived host cells. Those cells can contain endogenous retrovirus-like particles, which are potential source of contamination for recombinant biotherapeutics. Other sources of viral contamination could be environmental, personnel, and process-related factors such as raw materials used in media preparation.
Ensuring viral safety is crucial in bioprocesssing and is achieved by introducing several unit operations (e.g., low-pH viral inactivation and chromatography steps) to reduce viral load in a final drug substance. The process for such operations needs to be developed to establish reliable and robust viral clearance. To that end, extensive scaled-down wet-laboratory studies are needed. Although such validation studies are critical, their design and implementation are time and resource intensive. A digital twin could help pathogen-safety scientists to streamline the development of viral clearance processes.
Our approach to digital twins in pathogen safety is based on the development of multivariate data-driven models that are packaged in modeling applications. Those models are based on the implementation of machine learning (ML) algorithms used for recognizing patterns (unsupervised ML) and determining the relationship between inputs and outputs (supervised ML). In the case study below, three broad application categories were identified: contextualization of experimental results, predictive analytics, and what-if situations.
Contextualization of Experimental Results: Typical questions that scientists ask as they try to contextualize their experimental outcomes focus on how new experimental observations compare and contrast to historical expectation, whether there are groupings between runs, and which process parameters account for such data clusters. Such concerns can be addressed by applying principal component analysis (PCA) algorithms (5). PCA-based models can be used for pattern recognition and contextualization of experimental data to enhance process insights. That can be achieved by comparing new experimental designs against historical data (6).
Predictive Analytics: Questions about the extent of viral clearance for a given set of process parameter values can be addressed by conducting wet-laboratory experiments. Viral clearance across a unit operation typically is measured in terms of viral log-reduction factor (LRF). Predicting the LRF without running actual experiments can help expedite process development efforts at early stages. Supervised ML algorithms can leverage historical data for developing predictive models. It is worth noting that predictive models cannot replace actual experimentation fully. However, they can provide guidance for more efficient and targeted experimental designs. Panjwani et al. provide more details on ML-based predictive modeling (6).
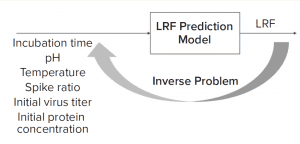
Figure 1: Solving an inverse problem is an approach to finding optimal process parameters given a target viral log-reduction factor (LRF).
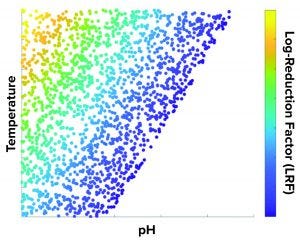
Figure 2: A temperature and pH design space is developed to achieve target LRF (numbers are not shown in the axes for confidentiality purposes).
What-If Scenario: Process development scientists often must determine the process parameter range to achieve a specific target viral reduction. Figure 1 shows one solution achieved by using an inverse problem in which the objective is to achieve target viral reduction and optimal process parameters are the outputs (Figure 1). Figure 2 shows the solution of one such inverse problem and illustrates a design space for two process parameters to achieve target LRF.
Process development scientists can use predictive models to contextualize historical data and test their hypotheses by first running those models and then performing only targeted wet-laboratory experiments. That can speed up process development for new protein drug candidates.
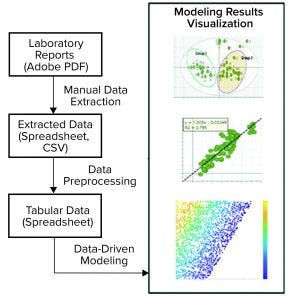
Figure 3: This pathogen safety model prototype consists of data extraction, processing, and modeling to achieve different model results visualization representations (CSV = comma-separated values).
Digital Twin Prototype: The application of multivariate data analysis (MVDA) and ML techniques to develop a useful model is just a small part of a modeling workflow. As Figure 3 shows, a typical workflow consists of data acquisition, preprocessing, model building and deployment, and visualization or reporting.
On-Premises Implementation: The first prototype of pathogen clearance and pathogen safety (PCPS) application was developed using multiple software and application-integrated on KNIME analytics platform (7). It started with manual data transcription from laboratory reports (Adobe PDF format) to flat files (Microsoft Excel spreadsheet or comma-separated values, CSV). To prepare data for ML application, flat file data were preprocessed and merged with publicly available flat file data using the KNIME analytics platform. The preprocessing involved standard steps such as data filtering, joining tables, handling missing information, and formatting. Preprocessed data then were fed to SIMCA MVDA software (from Sartorius Stedim Biotech) for pattern recognition and predictive modeling (8).
Model training was an iterative process, and communication with pathogen-safety scientists regarding model performance and interpretability was an essential part of this process. To solve the previously mentioned inverse problem (Figure 1) for determining the optimal range of process parameters, coefficients from a SIMCA model were exported in a flat file and used in the optimization framework developed using Python software packages. Given the target viral-reduction factor, the Python script produced optimal process parameter profiles (Figure 1). Finally, the developed model was implemented using an on-premises KNIME server, and an interactive web interface was created for the scientists to use that application.
Although, the prototype was a working solution for addressing pathogen-safety scientists’ concerns, it could not be scaled without addressing some limitations, the first of which was data acquisition. Viral safety data were transcribed manually from PDF reports, which made the process unscalable without increasing resources for data transcription. Manual data transcription also comes with the risk of transcription error. A machine-readable database could reduce that risk and simplify subsequent preprocessing steps.
SIMCA software facilitates exploration of widely used ML methods in the biopharmaceutical industry such as PCA and partial least squares (PLS) that balance interpretability and predictive ability. The software also comes with interactive visualization that makes early stage model development and exploration relatively easy. (Other advanced ML algorithms (9) such as random forest, decision trees, and neural networks are beyond the scope of this software.) In addition, the desktop-application nature of this proprietary software enables easy data exploration. However, the software has limitations when it comes to analyzing large data sets and implementing scalable models on cloud severs (mostly Linux based). With SIMCA software, as with other proprietary platforms, algorithm details are hidden from end users, so algorithm customization is not feasible.
Open-source software can address some of those constraints. For example, Python-based libraries such as scikit-learn, Keras, and TensorFlow models (10–12) are used widely in the technology industry for ML applications. Their open-source nature makes them transparent and customizable.
Cloud Implementation: To address the limitations described in the previous section, the next logical step was to use a cloud-based platform such as Amazon Web Services (AWS) for development and deployment of the pathogen-safety application. Traditionally, model development, implementation, and/or scale-up happen in silos. Lack of communication between development and deployment teams decreases overall efficiency and increases time for software-application release. DevOps (an approach combining software development and information technology operations) has emerged as an alternative approach for breaking down silos and ensuring continuous integration and continuous deployment (CI/CD) of software applications.
Implementing a CI/CD pipeline requires both a production and development environments. A development environment provides code developers with a safe space for improving a model or applications without worrying about the risk of unexpected impacts on the final application. A production environment is used for implementing the final application. Before pushing new changes to a final application in a production environment, code developers need to perform extensive testing. Code testing such as individual functional modules unit testing and complete integration testing can be automated to speed up integration and deployment of new changes to a final application in a production environment. That practice eliminates inadvertent disruption to a final application. Figure 4 shows the high-level architecture for a pathogen-safety application. That cloud architecture was setup to address the limitations described above with an on-premises solution. As discussed above, the data science environment serves as a sandbox for code development, whereas the production environment hosts the final application.
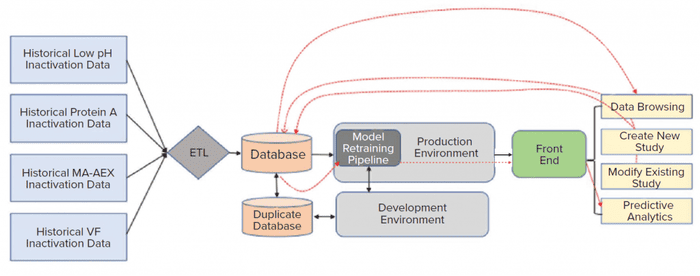
Figure 4: A cloud architecture for model development, deployment, and execution was designed to address the limitations of an on-premises system. ETL is extract transform load, VF is viral filtration, and MA-AEX is membrane adsorber anion-exchange chromatography.
AWS cloud data-storage services such as Amazon S3 and/or databases such as the MongoDB platform were used to collect data from historical and new studies. Historical data that reside in laboratory reports need to be processed first. The extract transform load (ETL) layer preprocesses those historical data. New laboratory-study data are entered directly through the front-end web interface and saved into appropriate databases. A direct data-entry option eliminates the need for manual data transcription and preprocessing. A single database for all conducted studies also greatly simplifies the data-browsing task. All modifications in existing study data can be made easily, and updated data can be stored in the MongoDB database. The three types of applications (contextualization of experimental results, predictive analytics, and what-if scenarios) can be executed by providing necessary user inputs through the front-end web interface. ML models used for such applications are stored in S3. Figure 5 provides a glimpse of the web-based front-end application. Django web framework was used to create the web interface.
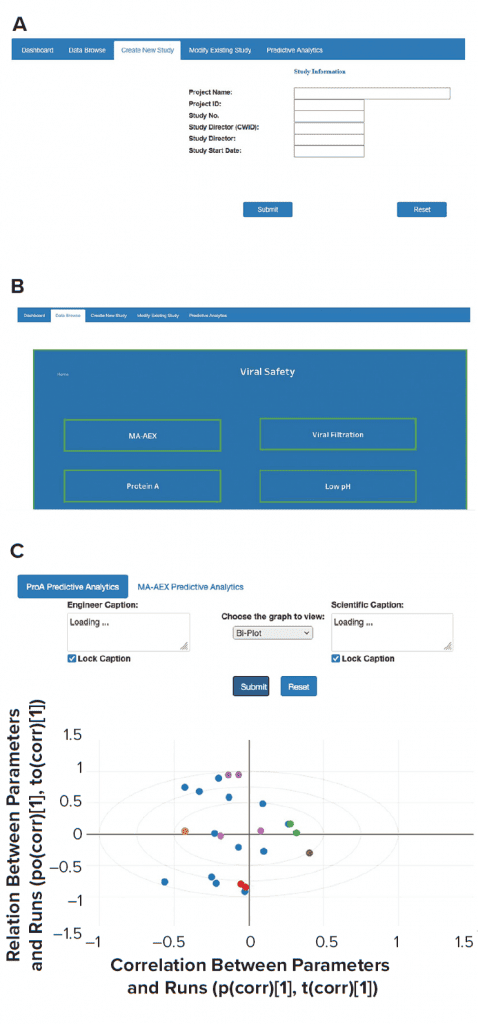
Figure 5: Screenshots were taken from front-end web interfaces for (a) creating a new study, (b) data browsing, and (c) performing analytics.
Learnings and Accomplishments: For this case study, a core cross-functional team was established. The team comprised experts from multiple departments and disciplines including information technology, laboratory automation, biological development, and data science. The project was defined as an exploratory innovation program, and the team was empowered and highly encouraged to explore and experiment. Key aspects contributing to the project’s success were establishing regular communication among the core team members and allowing time for ideation and frequent feedback on minimum viable products (MVPs). The team took a staged approach in that small packages of user requirements would be iterated until an MVP was developed before moving to the next unit operation. Multiple perspectives were needed to develop meaningfully a digital twin including: process information, understanding of metadata, establishing infrastructure and training and interpreting the models. The project had visibility and received support under the company’s digital transformation strategy, but it also had grass-roots elements in that the project team was empowered to define its own specific vision for the pathogen safety digital twin.
Significance of Digital Twin Platform
Overall, the development of digital twins for the PCPS case study and the cloud-based platform enables the reduction of the number of the wet-laboratory experiments needed. For example, such an approach can be used to evaluate new molecules without initial wet-laboratory experiments, thus mitigating manufacturability risk. Our approach also can support informed decisions regarding parameter evaluation for design-of-experiment (DoE) studies. Application of the models can drive improvements for downstream unit operations, enabling a holistic view of overall viral clearance for a particular process.
Adoption of a digital twin approach provides several advantages to a process development organization. In addition to improving efficiency (e.g., shortening times for data review of data and generating comprehensive visualizations and reports automatically), a shift to a data-centric view of process development can enable new paradigms.
Although proof-of-concept was conducted for a viral clearance process, a cloud-based architecture is general enough. Thus, it can be leveraged for implementing digital twins in other areas of biological development (e.g., cell culture) to support a holistic digital-aided design of manufacturing processes. A cloud-based architecture also contributes significantly to organizational efforts to leverage data science and digitalization in streamlining drug development processes. Most important, regulatory expectations can be met regarding the adoption of quality by design (QbD) methodologies that ensure safe and efficacious therapeutic molecules and processes.
Future Work
Although digital transformation and digital twins have different interpretations, they have promising applications in the bioprocessing industry. The case study herein showed an approach toward solving a particular business challenge, and a platform for expanding further-use cases and problem sets was established. The application of methodologies and approaches from other industries — specifically software development — has been highly successful and is now routinely used.
As mentioned above, the use of commercial software for ML-based applications helps in the proof-of-concept phase, but its use for cloud implementation could be challenging. Therefore, it has become important to adopt open-source technologies that can be tailored to fulfill the bioprocess industry’s digitalization needs. To that end, emphasis should be placed on in-house development of customized ML solutions for cloud deployment and exploration of new technologies for effective visualization.
Acknowledgments
We thank Xinkai Li, Karishma Desai, Ivan Cui, and Roger Canales for their contributions toward establishing the digital platform. We also acknowledge the entire pathogen safety team for its feedback during the platform-development phase.
References
1 Negri E, Fumagalli L, Macchi M. A Review of the Roles of Digital Twin in CPS-Based Production Systems. Procedia Manufact. 11, 2017: 939–948; https://doi.org/10.1016/j.promfg.2017.07.198.
2 Quality Guidelines. International Council on the Harmonisation of Technical Requirements for Pharmaceuticals for Human Use: Geneva, Switzerland; https://www.ich.org/page/quality-guidelines.
3 Guidance for Industry: PAT — A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance. US Food and Drug Administration: Silver Spring, MD, 2004; https://www.fda.gov/media/71012/download.
4 Guidance for Industry: Process Validation — General Principles and Practices. US Food and Drug Administration: Silver Spring, MD, 2011; https://www.fda.gov/files/drugs/published/Process-Validation–General-Principles-and-Practices.pdf.
5 Basilevsky AT. Statistical Factor Analysis and Related Methods: Theory and Applications. John Wiley & Sons: New York, NY, 2009: 197–181.
6 Panjwani S, et al. Application of Machine Learning Methods to Pathogen Safety Evaluation in Biological Manufacturing Processes. Biotechnol. Prog. 2021; 37(3) 2021: e3135; https://doi.org/10.1002/btpr.3135.
7 Berthold MR, et al. KNIME: The Konstanz Information Miner, Version 2.0 and Beyond. SIGKDD Explor Newsl. 11(1): 2009: 26–31; https://doi.org/10.1145/1656274.1656280.
8 SIMCA: Master Your Data. Sartorius Stedim Biotech; https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software/mvda-software/simca.
9 Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning, Data Mining, Inference, and Prediction, 2nd ed. Springer: Springer-Verlag New York, 2008.
10 Pedregosa F, et al. Scikit-Learn: Machine Learning in Python. JMLR 12, 2011: 2825–2830; https://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf.
11 Chollet F, et al. Keras: Deep Learning for Humans. GitHub 2015; https://github.com/fchollet/keras.
12 Abadi M, et al. Tensorflow: A System for Large-Scale Machine Learning. 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, 2–4 November 2016; https://www.usenix.org/system/files/conference/osdi16/osdi16-abadi.pdf.
Corresponding author Konstantinos Spetsieris is head of data science and statistics, Michal Mleczko is biotech digitalization lead, Shyam Panjwani is senior data scientist, Oliver Hesse is head of biotech data science and digitalization, all at Bayer U.S. LLC, Pharmaceuticals Division, 800 Dwight Way, Berkeley, CA 94710; 1-510-705-4783; [email protected]; http://www.bayer.com.
You May Also Like






