Maintaining Product Titer While Replacing Undefined Components in a CHO Culture SystemMaintaining Product Titer While Replacing Undefined Components in a CHO Culture System
September 1, 2009
Proteins, hydrolysates, and lysates of plant or yeast origin are commonly used in cell culture media for large-scale manufacturing processes for human biotherapeutics. Lot-to-lot variability in the composition of such constituents is well established and can affect multiple biological performance indicators. Our goal was to replace an undefined, protein-containing medium with a chemically defined medium (meaning the chemical structure and concentration for each component in a formulation is known). Such a formulation should be free of protein and any other components of direct animal origin (1). Adapting cells from medium containing hydrolysates and proteins to a chemically defined medium can be challenging because the cells must alter their gene transcription to adapt to the new conditions. However, the benefits of a defined, consistent, and predictable manufacturing process is often worth the investment of time and money. We developed a chemically defined medium and proceeded to optimize it for both growth and recombinant protein expression.
PRODUCT FOCUS: ANTIBODIES AND OTHER PRODUCTS OF ANIMAL CELL CULTURE
PROCESS FOCUS: PRODUCTION, PROCESS DEVELOPMENT
WHO SHOULD READ: PRODUCT AND PROCESS DEVELOPMENT SCIENTISTS, QA/QC, MANUFACTURING
KEYWORDS: SCALE-UP, OPTIMIZATION, MEDIA, FED-BATCH, DESIGN OF EXPERIMENTS, SERUM-FREE, HYDROLYSATES
LEVEL: INTERMEDIATE
Following adaptation to a chemically defined medium, we used design of experiment (DOE) methods to perform a media mixtures experiment, which revealed a mixture achieving higher viable cell density and protein titer compared with the original formulation. We followed the DOE study with fed-batch and bioreactor parameter optimization experiments that further improved viable cell density and protein titer. Finally, we analyzed the product expressed to determine what changes in protein quality had resulted from those optimization steps. The medium we developed produced higher viable cell densities and productivities than the original medium had, and the expressed protein had a higher degree of glycosylation with higher percentages of both G1F and G2F glycans.
Materials and Methods
Cells and Media: For this study we used a recombinant CHO cell line that had been engineered with CMC ICOS Biologics’ CHEF1 proprietary expression vector to express IgG4. The control medium was a CMC ICOS Biologics proprietary medium containing protein hydrolysates, recombinant protein, and high levels of glucose and buffering components (bicarbonate and phosphate). The test media consisted of four chemically defined formulations containing varying concentrations of glucose and buffering components (bicarbonate, HEPES, MOPS, and phosphate) but no protein or hydrolysates. Prototype Medium II resulted from the media mixtures DOE. Batch cultures were supplemented with 4-mM L-glutamine. We supplemented our fed-batch cultures with the following: CHO CD EfficientFeed A (Feed A), CHO CD EfficientFeed B (Feed B), and 200-mM L-glutamine supplements, all from Life Technologies (www.lifetechnologies.com).

Figure 1:
JUPITER IMAGES (WWW.JIUNLIMITED.COM)
Cell Maintenance and Adaptation: We grew shake-flask cultures (125-mL flasks with a 25-mL working volume for routine culture, 250-mL flasks with a 50-mL working volume for growth performance assays, and 500-mL flasks with a 100-mL working volume for scale-up culture) in a humidified 37°C incubator with 8% CO2 on an orbital platform rotating at 90 rpm. Bioreactor cultures were grown in either 5-L or 15-L (working volume) stirred-tank bioreactors from Applikon, Inc. (www.applikon.com), and we controlled pH (set-point 6.8), impeller speed, temperature (varied), and dissolved oxygen, DO (set point 60%).
We sequentially adapted cells from the control medium into four chemically defined media (Test media 1–4). The first step was to passage the cells from the control medium into various mixtures of it with each of the test media at ratios of 75/25, 50/50, 25/75, 10/90, and 5/95 test. We then passaged the cells from the media mixture containing the highest concentration of control into the next lower concentration of control, and so on, until they were fully adapted to the chemically defined medium. Cells were deemed ready for passaging when their doubling time reached ≤30 hours (data not shown). The seeding density for all experiments was 0.5 × 106 viable cells/mL (vc/mL).
Optimization: After DOE methods were used for the media mixtures studies (Figure 3), we analyzed growth and productivity data using Design Expert statistical software from Stat-Ease, Inc. (www.stat-ease.com). We supplemented Feeds A and B (15% at various intervals) in both shakers and bioreactors and evaluated temperature shifts (from 37 to 34°C and from 37 to 32°C) in the bioreactors.
Assays: For a growth performance assay (GPA), cells were seeded into and passaged in each test condition two times followed by the same test on passage 3. For the batch GPAs, cells were allowed to grow without refeeding until viability had dropped below 50%. For the fed-batch GPAs, cells were allowed to grow and were fed according to an appropriate feed schedule (described below) until viability had dropped below 50%. We initiated cell counts on either day 2 or day 3 after seeding, using a Cedex AS20 automated cell counter from Roche Innovatis AG (www.innovatis.com) to count and measure viability.
We quantitated immunoglobulin for the shaker-flask experiments on a protein G immunodetection sensor cartridge from Life Technologies and for the bioreactor experiments on a protein A HPLC system from Agilent Technologies (www.agilent.com). We examined glycosylation levels using a matrix-assisted laser-desorption ionized time-of-flight (MALDI-TOF) mass spectrometry system from Applied Biosystems (www.appliedbiosystems.com) and anion-exchange chromatography from Agilent. We elucidated the glycosylation levels by releasing N-glycans using PNGase F enzyme from Prozyme (www.prozyme.com), then derivatized the released glycans with 2-anthranilic acid (2 AA) from Sigma-Aldrich (www.sigmaaldrich.com) and measured their masses using MALDI-TOF or by separating them by anion-exchange chromatography.
Results
Adaptation: When we attempted direct adaptation from the control medium into test media, no cells survived. A sequential method provided the necessary environments for adaptation (Figure 1). The protocol that proved most effective involved passaging cells from the control medium into a mixture of 5% control medium and 95% Test 2 medium for one passage, then into 100% Test 2 medium. For passage 1 in the chemically defined medium (Test 2 medium), the cells appeared to lag (no growth, with viable cell densities remaining low until the final two days) as they adapted to grow in the new environment. By day 7, cells had doubled enough to allow their culture to be passaged. During the second passage, the cells again exhibited an extended lag, with growth beginning day 7 after seeding. By day 9, those cells had doubled enough to allow their culture to be passaged. During passage 3, the cells had doubled enough to allow their culture to be subcultured by day 3.
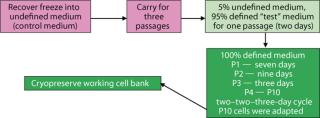
Figure 1: ()
For passages 1, 2, and 3, viable cell densities of 1.0–1.6 × 106 vc/mL were high enough for subculturing, even though such densities were lower than in subsequent passages. Starting with passage 4, we subcultured cells on a two-day, two-day, three-day cycle — and by passage 10, the cells had achieved viable densities of 2.0–4.0 × 106 vc/mL (equivalent to performance in the control medium). Those were used to seed subsequent experiments.
Growth Performance Assays (GPA): Once the cells were fully adapted to Test 2 medium, we performed a baseline growth curve. Test 2 medium (6.0 ± 0.6 × 106 vc/mL peak cell density, 259 ± 8 mg/L titer) performed equivalent to the control medium with hydrolysate lot B (5.1 ± 0.02 × 106 vc/mL peak cell density, 282 ± 8 mg/L titer), but not as well as the control medium with hydrolysate lot A (8.7 ± 0.6 × 106 vc/mL peak cell density, 620 ± 3.6 mg/L titer) (Figure 2).
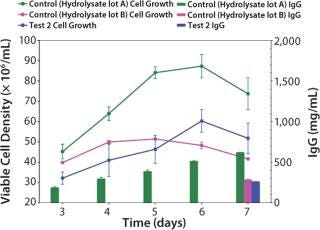
Figure 2: ()
We designed a media mixtures DOE (Table 1) and carried it out using shake flasks. Figure 3 presents the four test media (Test 1–4 media) and the two best-performing mixtures (A and B) based on antibody titer from this GPA. Medium A is a mixture of Test Media 1, 2, 3, and 4; Medium B is a mixture of Test Media 1, 2, and 4. Medium A grew cells to a higher peak density (8.4 ± 0.2 × 106 vc/mL) than did Medium B (6.7 ± 0.3 × 106 vc/mL) with equivalent production (~420 ± 18.7 mg/L). And Medium B supported a higher specific productivity (12.5 pg/cell/day) than did Medium A (10.3 pg/cell/day).
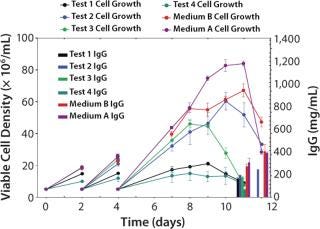
Figure 3: ()
Table 1: Media mixtures DOE design layout

Table 1:
Scaling our test system up into a 5-L batch bioreactor, we compared Medium A with an undefined control for its ability to support cell growth and productivity. The results (Figure 4) demonstrate that Medium A reached a similar level of productivity to the control, so it is possible to remove all undefined components without a loss of performance. A benefit was that a peak titer level was reached about four days earlier in Medium A than in the control medium. Amino-acid analysis of spent media showed that several components were depleted and may have become rate limiting (data not shown).
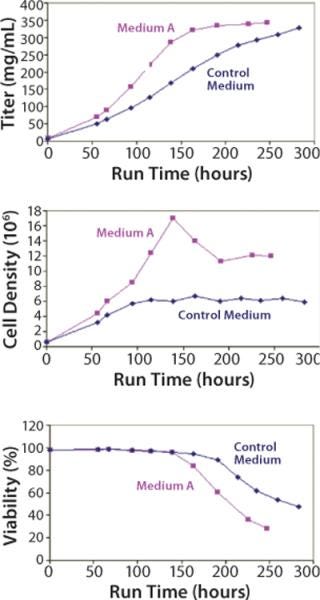
Figure 4: ()
With the successful creation of a chemically defined growth medium (Medium A) that supported both growth and productivity comparable to the undefined control medium, our next step was to create a synergistic, chemically defined feed supplement. We performed fed-batch shake flask experiments (Figure 5) comparing Medium A with feed (a new process) to the control medium with feed (the original process). Based on previous experience, we fed Medium A with a 1:1 mixture of Feeds A and B (15% by volume) on days 0, 3, 6, and 9. We fed the control medium with a control feed (containing hydrolysate) on days 3, 4, 5, 6, and 7, which is the CMC ICOS Biologics standard protocol. The chemically defined feed supplement performed better (650 mg/L) than the undefined feed supplement (380 mg/L). In addition, use of the chemically defined feed supplement increased product titer over batch mode (320 mg/L, Figure 3).

Figures 5A and B: ()
We then scaled up our chemically defined system into 15-L bioreactors. Cells were grown in Medium A and fed a 1:1 mixture of Feeds A and B (15% by volume) using three different feed schedules along with a temperature shift from 37°C to either 34 or 32°C on day 3 (Figure 6). We tested three feed schedules: feed 1 (feed added on days 0, 3, 6, and 9), feed 2 (feed added on days 0, 2, 4, and 6), and feed 3 (feed added on days 3, 6, 9, and 12). When the concentration of L-glutamine fell below 1.5 mM, we added it in bolus form to bring the concentration back to 4 mM. Peak viable cell densities for the temperature shift from 37 to 34°C were between 8 × 106 and 10 × 106, depending on the feed schedule. Peak viable cell densities for the temperature shift from 37 to 32°C were ~7.5 × 106. Protein titers ranged 700–943 mg/L.
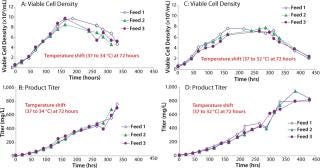
Figures 6, A–D: ()
Protein Quality: To check product glycosylation, we performed fed-batch shake-flask experiments comparing antibodies produced in the hydrolysate-containing culture system with those expressed in the chemically defined culture system (Figure 7). Over 90% of the total HPLC peak area was assigned as biantennary, core-fucosylated N-linked glycans with 0, 1, or 2 terminal galactose residues (designated G0F, G1F. and G2F, respectively) by MALDI-TOF (data not shown). Peak areas measured by high-performance liquid chromatography (HPLC) showed that antibody produced in the chemically defined culture system had higher percentages of the G1F and G2F glycoforms (40% G0F, 40% G1F, and 15% G2F) than that produced in the undefined culture system (55% G0F, 30% G1F, and 7% G2F). A small peak was observed in the MALDI spectrum corresponding to a high-mannose glycan (Hex5HexNAc2), but it was not quantified by HPLC, and no masses corresponding to Gal-Gal species (e.g., G2F-Gal) were observed in either sample.
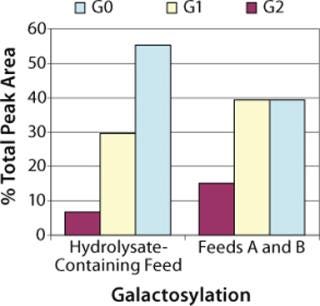
Figure 7. ()
Discussion and Conclusions
Using statistically designed experimental methods, we developed a chemically defined culture medium that supports growth and IgG production. In fed-batch bioreactors using the optimized growth medium (Medium A), Feeds A and B, and a temperature shift from 37 to 32°C, we successfully obtained a production level of ~940 mg/L (Figure 6D). Our chemically defined culture system supports production at a level higher than the protein-containing, undefined culture system — even when supplemented with the higher performing lot of hydrolysates (~600 mg/L) (Figure 2).
The cell line we used was previously shown to reach titers in the range of 500 mg/L to >1 gram/L when used in a fed-batch process containing recombinant protein and plant hydrolysate. The large range in titer has been attributed to lot-to-lot variability in the hydrolysate supplement. Our chemically defined medium should eliminate such process variability and lead to a more predictable and robust manufacturing process. Additionally, elimination of undefined bulk from the medium should make it possible to streamline downstream processing and make it more cost effective. This may also lead to recovery of a higher percentage of purified product.
Further optimization of our feed strategy may improve performance. Feed strategy development can be accomplished by evaluating spent media analysis results and then modifying the feed formulation by reducing the concentrations of components that are not being used — and increasing the concentration of the components that are being depleted. As cell densities increase, some components (e.g., magnesium, potassium, and phosphate) may become rate limiting because they are being taken up into cells and are therefore unavailable in the medium (2). Such components may need to be added to the feed.
Balancing nutrient composition throughout a culture cycle is difficult but necessary for optimum efficiency. Our study shows that the best culture condition tested was when CHO cells were grown in a fed-batch bioreactor with a temperature shift from 37 to 32°C. However, further optimization of the bioreactor operating parameters could yield improved growth and protein titers. Changing the pH set point or using a pH shift may improve performance. Additionally, modifying the sparge rate and/or impellor configuration/speed (particularly as the system is scaled up into production vessels) could improve performance by clearing CO2 and/or improving the availability of oxygen to the cells (3,4).
The product displayed higher percentages of G1F and G2F glycans in our chemically defined fed-batch culture system than in the undefined culture system (although it is not clear from existing literature whether more or less galactosylation is generally desirable). Because we conducted our glycosylation studies on protein from shake flasks, the glycosylation pattern of protein produced in the more controlled environment of a bioreactor still needs to be investigated. The outcome of such a study may require reevaluation of the optimization steps and goals. Optimizing a feed strategy to reduce ammonium concentration (data not shown) may also help improve product titer and alter the glycosylation pattern.
Protein hydrolysates have been used for many years to improve growth and production, but they have many potentially negative attributes (5). Here, replacing hydrolysates with a chemically defined environment has helped cells produce a higher concentration of protein. The expressed product has a higher degree of galactosylation with more terminal galactose residues present on the antibody glycans. Moving from a hydrolysate- and protein-containing medium to a chemically defined formulation reduced lot-to-lot variability attributed to those undefined medium components. Because we reduced lot-to-lot variability, this biotherapeutic manufacturing process will be more robust and predictable. Understanding the nature and quality of raw materials used in a biomanufacturing process — and their impact on quality of the expressed product — are key parts of FDA’s process analytical technology (PAT) initiative (6). Using chemically defined culture media and feeds, companies will be in a much better position to fully capitalize on the opportunities of that initiative.
REFERENCES
1.) Gorfien, S. Butler, M 2007.Chapter 5: Cell Culture Media Development — Customization of Animal Origin-Free Components and SupplementsCell Culture and Upstream Processing, Garland Science Publishing, New York, NY, Oxford:81-98.
2.) Wlaschen, KF, and W-S. Hu. 2006. Fedbatch Culture and Dynamic Nutient Feeding. Adv. Biochem. Engin./Biotechnol. 101:43-74.
3.) Jan, DCH. 1997. The Effect of Dissolved Oxygen on the Metabolic Profile of a Murine Hybridoma Grown in Serum-Free Medium in Continuous Culture. Biotechnol. Bioeng. 54:153-163.
4.) Restelli, V. 2006. The Effect of Dissolved Oxygen on the Production and the Glycosylation Profile of Recombinant Human Erythropoietin Produced from CHO Cells. Biotechnol. Bioeng. 94:481-494.
5.) Lobo-Alfonso, J, P Price, and D. Jayme Pasupuleti, VK. 2008.Benefits and Limitations of Protein Hydrolysates As Components of Serum-Free Media for Animal Cell CultureProtein Hydrolysates in Biotechnology, Springer Publishing Company, New York.
6.) CDER/CVM/ORA September 2004. Guidance for Industry: PAT — A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance, US Food and Drug Administration, Rockville.
You May Also Like






