Automation of MicrobioreactorsAutomation of Microbioreactors
Current methodologies in genetics and microbiology enable researchers to influence metabolic pathways of microbial cells in many directions. Beside the academic interest in investigating fundamental functions in metabolic pathways, commercial production of valuable compounds by microbial hosts is state of the art. For example, such products include enzymes (lipases, proteases, phytases), therapeutic agents (insulin, antibodies), bulk chemicals (lysine, glutamate, citric acid), or the microbial cells themselves (used in brewing or milk processing), with therapeutic agents probably the fastest growing market.
Reducing cost is a driving interest in commercial bioprocesses. Therefore, genetic manipulations can provoke the transformation of low- into high-producing strains. Such work can generate a huge number of candidates to be screened for higher productivity, as shown by Brockmeier et al., who generated a library of 148 signal peptides in Bacillus subtilis to enhance secretion of heterologous expressed proteins (1). It was not predictable which combination of signal peptide and recombinant enzyme resulted in high secretion efficacy. That highlights the need for automated and parallel screening systems when developing bioprocesses from scratch.
PRODUCT FOCUS: RECOMBINANT PROTEINS
PROCESS FOCUS: PRODUCTION
WHO SHOULDREAD: PROCESS ENGINEERS, FERMENTATION SCIENTISTS, MICROBIOLOGISTS
KEYWORDS: MICROBIOREACTOR, PROCESS MONITORING, PROCESS OPTIMIZATION, PROCESS DEVELOPMENT, LAB AUTOMATION
LEVEL: INTERMEDIATE
In addition, process productivity strongly depends on cultivation conditions, which may be different for each strain or construct to be screened. This was shown for a library of 444 clones of Hansenula polymorpha expressing GFP as model protein by Scheidle et al. (2). In that library, the expression cassette was integrated into the genome at different loci and under control of either the FMD-or MOX-promotor. The authors cultivated clones in fed-batch mode (realized with slow-release of glucose) and batch mode using glucose or glycerol as the sole carbon source. In addition, those three modes were conducted in two different media backgrounds. The resulting clone rankings were different for all cultivation setups, and the authors concluded that clone screening should be performed under similar process conditions as in production scale. Thus, small-scale and parallel-operated bioreactor systems are the cultivation vessels of choice for handling the increased number of experiments to surely identify the most promising combination of production strain and bioprocess mode. Very often, microbioreactors are used to perform such studies (3).
Because cultivations at laboratory and production scales are commonly run in fed-batch mode, it is desirable to translate this into the microliter scale of microbioreactors to match conditions. Fed-batch process control (that is, the feeding of nutritional compounds to a culture over time) is generally known to be beneficial in terms of higher product titers and productivity. By contrast with common fermentor systems, in which feeding solutions are fed into a vessel with pumping systems (because of the large volumes applied), this was not possible at microliter scale until now. Addition of very small volumes (nL to μL) has to be realized to scaled-down common fed-batch bioprocesses.
Herein, we present the RoboLector microbioreactor system, which performs fed-batch cultivations in microliter scale. We discuss hardware configurations to match different needs in upstream bioprocess development and demonstrate its feasibility on the basis of two application examples. The system can be applied in investigations described in literature, so it enables greater degrees of freedom for further improvement and understanding of those works.
Configuration and Operating Mode
The RoboLector microbioreactor system integrates the BioLector system into a liquid-handling robot, described for the first time by Huber, et al. (4). The BioLector system provides relevant online monitored fermentation data from up to 48 or 96 parallel cultivations through microplates. Those data are measured repeatedly within 5–15 minutes for each parallel cultivation. Based on the online monitored data (and on other predefined events) each single cultivation can be manipulated by the liquid-handling robot individually. That includes feeding of nutrients or inducers, addition of basic or acidic solutions for pH-adjustment, and sampling.
Photo 1 and Figure 1 depict the RoboLector microbioreactor system. Figure 1 visualizes the controlling loop of an experiment as follows: A user defines an experiment and triggers for the operation of the liquid-handling unit. For example, if a rise in dissolved oxygen (DO) signal >80% of air saturation is detected, then some feed solution is added or if pH drops below 6.9, then basic solution is added. The BioLector device monitors fermentation parameters such as DO or pH continuously. Based on those data, the supervising software orders the liquid-handling unit to act on the cultivations.
Photo 1:
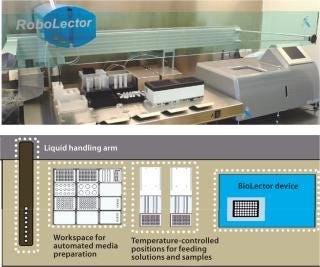
Photo 1: ()
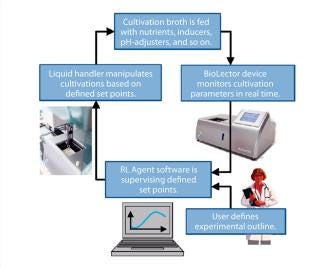
Figure 1: ()
Photo 1 shows the RoboLector XL system, with details of its components below. The system contains a BioLector device, a liquid-handling arm, positions for addition of solutions, and samples taken from individual cultures. All positions are temperature controlled and allow samples to cool or freeze to stop metabolic activity. Also, a workspace for automated media preparation is implemented on the robotic deck. Application of design of experiments (DoE) can result in complex media formulations that the robotic system can prepare easily and autonomously.
Third-party devices can be integrated to expand the possibilities of this system. One customized RoboLector system was described with an integrated centrifuge for cell separation and a microplate reader to conduct enzymatic assays for determination of recombinant target proteins (5). The RoboLector is not preassigned to a specific manufacturer’s liquid-handling unit, so maximum flexibility is achieved to satisfy specific requirements or tasks in bioprocess development.
Fed-Batch Cultivation of Escherichia coli
E. coli is one of the workhorses in recombinant protein expression known for low doubling times a
nd high protein synthesis rates. In classical E. coli fed-batch cultivations using stirred-tank reactors, feeding solutions normally contain a carbon source, ammonium, and magnesium to match the nutritional requirements.
Figure 2 depicts two fed-batch cultivations with the RoboLector system using different compositions of those nutrients. Sodium hydroxide was included to stabilize the pH value during cultivation. After batch growth for approximately six hours on the initially included carbon source (10 g/L glycerol), the repeated addition of the feeding solutions was triggered by online monitored DO measurements. Rising DO levels indicated depletion of carbon source and thus, the optimal time point for addition of a new portion of carbon source.
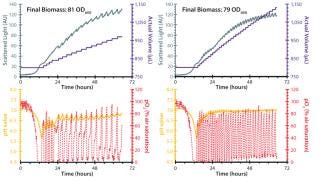
Figure 2: ()
It is notable that both feeding strategies promoted growth to high cell densities of ~80 OD600 in combination with stabilization of the pH value in a favorable range for growth of E. coli. During initial batch growth, pH drops below 6.0 because no pH-adjusting agents are added until feeding starts. During the feeding phase, pH stabilizes between 6.5 and 7.0, caused by the included sodium hydroxide in the feeding solution. The nutritional solution containing 200 g/L glycerol as a carbon source resulted in smaller intervals of feeding compared with the 400 g/L glycerol solution. Therefore, the volume increased slower for the latter.
With the same biomass concentration for both setups, but different resulting reaction volumes, a higher absolute amount of biomass is generated (biomass concentration multiplied with reaction volume) when applying 400 g/L glycerol in the feeding solution than when applying 200 g/L. That may be a concern in scale-up studies, because higher volumes need bigger reactors and more energy for mixing, heating, and cooling. With such high cell density cultivation (HCDC) setups for E. coli, the RoboLector system proves to be a precise and very efficient tool for fast bioprocess development with microbial cells.
Enhancing Productivity of a Pichia Pastoris Fed-Batch Process Using DoE
During bioprocessing, secreted proteins are very often confronted with proteolysis and thus continuously degraded. It is very common practice, therefore, to take samples from a fermentation broth to generate a time course (kinetics) of product concentration and activity. We transferred this process to the RoboLector system.
For our example, we used automated sampling to obtain a time course of the activity of the recombinant enzyme. In a process optimization study using DoE, those samples were analyzed at-line to determine the optimal cultivation setup to reach maximum activity.
Figure 3 (LEFT) shows detailed kinetics of a P. pastoris fed-batch cultivation carried out with the RoboLector system. The online monitored data, the concentration of the secretory expressed enzyme of interest, and corresponding space-time-yield are plotted. The amount of recombinant protein was determined at-line from automatically drawn samples of 20 μL. By contrast with dosing nutrient solutions for sampling, the robotic tips immerse in the fermentation broth without interruption of shaking (Figure 3, RIGHT). Doing so provides a representative sample of the cell suspension, preventing artifacts of sedimentation without shaking.
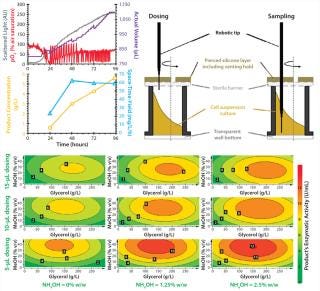
Figure 3: ()
In our example (compare Figure 3, LEFT), product concentration continuously increases until the end of cultivation and thus, space-time-yield does not drop from the maximum value reached after 48 h. This was not the case for other strains from the screened library (data not shown).
For this clone, we performed further investigations to enhance productivity. Commercial DoE software planned the experimental outline autonomously according to input factors. These factors were as follows: composition of feeding solution (variations in concentrations of methanol, glycerol, and ammonium hydroxide) as well as applied dosing volume. Because of the possibility to conduct up to 48 parallel fed-batch cultivations, a full-factorial design plan (24 + 3 = 19 variations in total) could be carried out in one experiment.
Using all cultivation results according to the conducted DoE plan, a contour plot was automatically generated (Figure 3, BOTTOM). It summarizes the maximum activity of the recombinant enzyme with respect to the different applied compositions of feeding solutions and dosing volumes. With this plot, optimal conditions can be easily visualized. In our case, the highest activity is located at the bottom-right part of the contour plot. To be precise, a maximum activity of the recombinant product (and thus, concentration) could be obtained for the following combination of factors: dosing volume = 5 μL and a feed composition of 35% v/v methanol, 150 g/L glycerol, and 2.5% w/w ammonium hydroxide.
Those results can be used to curtail the factors for further investigations. In general, at least two successive runs are recommended when DoE is applied. The first is for identifying factors of influence and their interactions. The second is for fine-tuning those identified factors to enhance productivity.
Outlook
In most fermentation laboratories, bioprocess development is still performed using stirred-tank bioreactors at 0.5 L to 10 L scale small-scale technologies are lacking or unavailable. The increasing number of projects and clones to be screened has urged researchers to find more efficient technologies and adapt their laboratories to high-throughput experimentation. Our high-throughput fermentation system overcomes most current limitations of small-scale cultivation systems and performs 48 parallel fed-batch bioprocesses at 1-mL scale.
The system’s fed-batch capability allows researchers to expand their repertoire to maximize bioprocessing productivity, as demonstrated by application examples for E. coli and P. pastoris. We conclude that sophisticated optimization procedures for E. coli — like those described by Huber et al. (6) and Islam et al. (7) — will clearly benefit from the possibility to feed nutrients based on online monitored fermentation data. And we assume that the impressive work by Studier (8) to develop auto-inducing media could have been a target for the RoboLector system (e.g., to optimize auto-inducing feeding solutions that may be superior to classical induction with ITPG in T7-based expression for E. coli).
Scheidle et al. (2) demonstrated that clones selected under batch-screening conditions are probably not the best producers at final production scale (fed-batch mode) because of their different operation modes. The authors applied polymer reservoirs with a constant glucose release to apply fed-batch conditions and were able to select completely different clones in fed-batch than in batch conditions. Because of the limited flexibility of their feeding technology, they were unable to investigate the effects of carbon sources other than glucose, especially a fed-batch process based on glycerol.
The RoboLector microbioreactor system can overcome such limitations. It is possible to perform several controlled fed-batch cultivations simultaneously. So upstream bioprocess development for enhancing productivity can be conducted in a short time frame. By performing cultivations at microliter scale close to production conditions (e.g., in fed-batch mode), researchers can now identify such high-producing clones more safely. That prevents surprises of low-performing clones in later bioprocess development stages and streamlines bioprocess development procedures to reduce time to market.
The high-throughput capacities of this system facilitates application of DoE software tools. Now it is possible to investigate the interaction of different producer strains in combination with different fermentation conditions at once and not in sequence as it is performed nowadays. We demonstrated DoE application with a P. pastoris fed-batch process. A full-factorial design plan performed automatically, and a contour plot highlighted the best process conditions for the highest product concentration/activity.
The RoboLector microbioreactor system can perform 48 fed-batch fermentations in parallel automatically. Standard fed-batch processes were transferred to the automated system and experiments planned with DoE were performed successfully. We believe that the majority of experiments performed using shake flasks or stirred-tank bioreactors can be easily down-scaled and transferred to the RoboLector microbioreactor system, so experiments can be performed automatically and at higher throughput to accelerate bioprocess development. In the near future, stirred-tank bioreactor runs should have to be conducted only for confirmation runs or material production.
About the Author
Author Details
Johannes Hemmerich is a scientific officer, and corresponding author Frank Kensy is the managing director at m2p-labs GmbH, Arnold-Sommerfeld-Ring 2, 52499 Baesweiler, Germany; 49-2401-805331; fax 49-2401- 805333; [email protected]; www.m2p-labs.com.
REFERENCES
1.) Brockmeier, U. 2006. Systematic Screening of All Signal Peptides from Bacillus subtilis. A Powerful Strategy in Optimizing Heterologous Protein Secretion in Gram- Positive Bacteria. J. Mol. Biol. 362:393-402.
2.) Scheidle, M. 2010. High-Throughput Screening of Hansenula polymorpha Clones in the Batch compared with the Controlled- Release Fed-Batch Mode on a Small Scale. FEMS Yeast Res. 10:83-92.
3.) Marques, MPC, JMS Cabral, and P Fernandes. 2009. High-Throughput in Biotechnology: From Shake-Flasks to Fully Instrumented Microfermentors. Recent Pat. Biotechnol. 3:124-140.
4.) Huber, R. 2009. RoboLector: A Novel Platform for Automated High-Throughput Cultivations in Microtiter Plates with High Information Content. Microb. Cell Fact. 8:42.
5.) Rohe, P. 2012. An Automated Workflow for Enhancing Microbial Bioprocess Optimization on a Novel Microbioreactor Platform. Microb. Cell Fact. 11:144.
6.) Huber, R. 2011. Utilizing High- Throughput Experimentation to Enhance Specific Productivity of an E. coli T7 Expression System by Phosphate Limitation. BMC Biotechnol. 11:22.
7.) Islam, RS. 2007. Framework for the Rapid Optimization of Soluble Protein Expression in Escherichia coli Combining Microscale Experiments and Statistical Experimental Design. Biotechnol. Prog. 23:785-793.
8.) Studier, FW 2005. Protein Production By Auto-Induction in High-Density Shaking Cultures. Protein Express Purif. 41:207-234.
You May Also Like





