Efficient Development of Stable High-Titer Cell Lines for Biopharmaceutical ManufacturingEfficient Development of Stable High-Titer Cell Lines for Biopharmaceutical Manufacturing
Commercial manufacturing of therapeutic monoclonal antibodies (MAbs) commonly uses mammalian cells to generate large quantities of a drug. Identifying cell lines that stably produce high protein titers is, therefore, a critical part of biopharmaceutical development. Unfortunately, identifying suitable cell lines is traditionally a time-consuming, labor-intensive process. That’s because their productivity and stability can vary enormously, so large numbers of clones must be screened to find those with both the highest yield and a desired level of product quality (1).
Cell-line development is unsurprisingly seen as a major bottleneck in biopharmaceutical development (2,3). With companies facing pressure to reduce costs and shorten development times, techniques to improve the efficiency of this process are in demand. Several methods have been developed, including fluorescence-activated cell sorting (FACS) such as the BD FACS system from BD Biosciences (www.bdbiosciences.com) and the ClonePix FL system from Genetix, Ltd. (www.genetix.com) (4) to speed up cell-line selection and identify high-producing cell lines at an early stage in cell-line development (2,3,4,5). However, limited progress has been made in reducing the manpower costs involved in expanding several clones in cell-line development. We present an automated approach for cell-line development based on the Cello robotic system from The Automation Partnership (www.autoprt.co.uk). We believe it offers significant advantages over manual techniques.
PRODUCT FOCUS: BIOPHARMACEUTICALS
PROCESS FOCUS: PRODUCTION
WHO SHOULD READ: R&D AND PROCESS DEVELOPMENT
KEYWORDS: CELL-LINE DEVELOPMENT, DRUG DISCOVERY, AUTOMATION, CHO CELLS, ROBOTICS
LEVEL: INTERMEDIATE
Problems with Traditional Approaches
The most common mammalian cell lines used to manufacture commercial quantities of therapeutic antibodies are derived from Chinese hamster ovary (CHO) cells. Traditionally gene expression systems based on dehydrofolate reductase (DHFR) and the more recent glutamine synthetase (GS) from Lonza (www.lonza.com) have been used for development of cell lines suitable for commercial manufacturing.

In traditional CHO-based expression systems, plasmid DNA is integrated into host-cell chromosomes during transfection. The plasmids contain the genes required for cells to manufacture a therapeutic antibody and selection markers required to identify those cells that do express the desired protein. However, productivity of cell lines generated using these expression systems varies enormously because plasmid DNA is randomly integrated, and drug yield depends on the integration site (6).
Generally, few transfected CHO cells will be high producers, and the number of high-producing clones detected depends on the number screened (2). Screening many clones and expansion of selected cell cultures to larger scales is time-consuming and labor-intensive. In addition, tracking documentation becomes a challenge with large numbers of clones. Colony screening is traditionally done manually using microscopy. It is followed by expansion of a large number (200–1,000) of clones into static cultures that are then screened for IgG titers in batch culture. Selected cell lines are expanded into a suspension culture for further evaluation using batch and fed-batch cultures grown in shake flasks and bioreactors.
Cell-line selection techniques using fluorescence technology were developed to be a less labor-intensive alternative to traditional screening for colonies and productivity. Commonly, such systems select high-producing cells by imaging fluorescently-tagged antibodies that bind to a drug protein. However, these techniques are applicable only during early selection. Expansion of static cultures — and subsequent evaluations in suspension cultures — still have to be performed manually or using additional equipment. We chose instead to study how that process could be automated to make cell-line development more efficient and less labor-intensive. To further cut down required man-hours, this automation approach can be combined with fluorescence techniques as well. Several other expression systems have been developed in recent decades for improvement of titer or shortened timelines (7).
Benefits of Automation
An automated concept for developing high-producing cell lines for biopharmaceutical production has now been successfully demonstrated (8). The approach uses a Cello robotic system, which can operate 24 hours a day and performs optical clone screening using automated microscopy and scale-up of static cultures. This system enables high-throughput, off-line screening of IgG titer in batch cultures at different stages during scale-up. Its software records all process steps and associated data in a relational database together with clone images to facilitate detailed documentation of the selection process.
We have shown that the automated approach generates cell lines of equal quality to those produced by traditional manual selection procedures. Throughput of cell-line generation is increased while maintaining or even reducing cell-line development costs. Our models show that up to three times as many cell lines can be screened by the same number of staff using this automated technique while continuing to screen large numbers of clones. Large screens also provide a higher chance of finding high-producing cell lines. Furthermore, automatic tracking of clones through the expansion process gives an added advantage of providing proper documentation for a strictly regulated environment.
An Automated Platform
Figure 1 shows a Cello system and its configuration: a plate-handling robot, up to three incubators, a cold storage unit, two liquid handling units with pipetting robots, and a high-resolution MAIA Scientific microscope. The high-resolution microscope is used for clone detection by imaging cell cultures in multiwell plates (Photo 1). The system records all process steps carried out on each individual well along with other associated data. That way, each clone can be traced easily, and detailed records are kept for regulatory purposes (8).
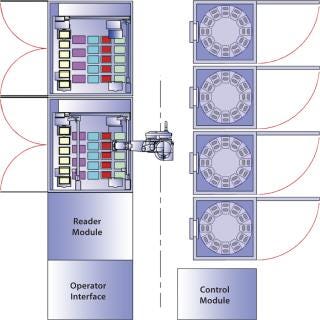
Figure 1: ()
Photo 1:

Photo 1: ()
Cello software can schedule multiple cell-line development experiments in parallel. Following a predefined procedure, the robot carries out all process steps involved in screening and expansion of static cultures, then selects clones for further analysis using assay data loaded into the system. As an option, single-cell cloning can be performed with a limiting dilution process.
Figure 2 shows a schematic of the cell-line development process we used. The colonies are assessed just as in the traditional manual procedure except that the procedure is highly automated. Following transfection, 96-well culture plates are placed in Cello incubators. The system automatically detects single colonies growing in those plates over a period of two to four weeks. The colonies could be prescreened off-line to remove low- or nonproducers based on an enzyme-linked immunosorbent assay (ELISA), and the remaining colonies then expanded to 24-well plates.

Figure 2: ()
From that stage on, an automated off-line protein A high-performance liquid chromatography (HPLC) analytical method is used to select 20–30% of the best-producing clones, which are then expanded into six-well plates. About 15–20% of the remaining clones are then selected for further evaluation in shake flasks outside the Cello system (8). For measuring protein expression without a protein A HPLC technique, a high-throughput immunoassay from Gyros AB (www.gyros.com) is available for screening (9).
Demonstrated Success
Both Lonza’s GS expression system and the DHFR-based OptiCHO expression system from Invitrogen (www.invitrogen.com) have been successfully adapted to the automated platform. A recent study using the GS system compared the quality of cell lines produced using the automated platform with those produced using a traditional manual procedure. As Figure 3 shows, monoclonal antibody concentrations produced by selected clones were comparable for both procedures (8).
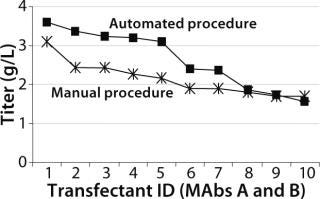
Figure 3: ()
Similar numbers were screened in both the manual (800) and automated (1,000) processes. The manual method was performed as two experiments because it was difficult to handle the same numbers the robot could. The automated procedure for screening cell lines was as described above. In the manual procedure, colonies were visually detected in 96-well plates three to five weeks after transfection. Single colonies were selected for ELISA, then manually expanded from 96-well to 24- and six-well plates. Based on IgG titers in the HPLC primary screen, selected clones were transferred to shake flasks (8).
Cell lines selected by both procedures were expanded to shake flasks. When the cell cultures had entered their exponential growth phase, they were evaluated in a batch process at 50-mL cultivation volume in 250-mL shake flasks.
The best performers were further examined during a 14-day fed-batch process. After harvest, their IgG concentration was measured and the top cell line studied further using a generic fed-batch process in stirred-tank bioreactors. The top-performing cell line from the manual procedure was cultivated to 20-L volume in a stainless steel stirred-tank bioreactor. The best cell line from the automated procedure was cultured to 25 L in a single-use bioreactor.
The IgG titer distribution of the batch cultures in 24-well plates and shake-flask batch cultures were similar for both processes. In addition, IgG titers for the top cell lines in both processes was comparably high. The automated procedure maintained cell growth and survival during cell-line development (8), which is a general prerequisite for the routine application of automation in cell line development. Similar expression levels and growth curves were found during a further experiment using the automated procedure on clones expressing a human IgG4 antibody.
Challenges and Future Work
The study described here shows that an automated procedure for cell-line development can generate high-producing cell lines while reducing costs and manpower requirements. The robotic system allows both screening of many cell lines and simultaneous performance of several projects (8). Two different expression systems have been successfully implemented, with the OptiCHO system being most recent. Our main focus for ongoing work is on optimizing parameters in the automated procedure to shorten time lines and further increase expression levels as well as the number of high-producing cell lines achievable with the OptiCHO system.
Future improvements will focus on further reducing the manual handling required and increasing the capacity of the robotic system. This may involve reducing the number of culture plates handled in each project by removing low-producing transfectants at an earlier stage. However, it is important to be cautious. We have found it hard to predict from early stage results which clones will turn into high-producing cell lines in bioreactor fed-batch cultures (8).
REFERENCES
1.) Birch, JR, and AJ. Racher. 2006. Antibody Production. Adv. Drug. Deliv. Rev. 58:671-685.
2.) Carroll, S, and M. Al-Rubeai. 2004. Selection of High-Producing Cell lines Using Flow Cytometry and Cell Sorting. Expert Opin. Biol. Ther. 4:1821-1829.
3.) Sleiman, RJ. 2008. Accelerated Cell Line Development Using Two-Color Fluorescence Activated Cell Sorting to Select Highly Expressing Antibody-Producing Clones. Biotechnol. Bioeng. 99:578-587.
4.) Mann, C. 2006. Rapid and Efficient Screening and Isolation of Antibody- and Biopharmaceutical-Secreting Cell Lines. Nature Meth. 1:SS4-SS5.
5.) Brezinsky, SCG. 2003. A Simple Method for Enriching Populations of Transfected CHO Cells for Cells of Higher Specific Productivity. J. Immunol. Meth. 277:141-155.
6.) Wurm, FM. 2004. Production of Recombinant Protein Therapeutics in Cultivated Mammalian Cells. Nature Biotechnol. 22:1393-1398.
7.) Clarke, HRG, and BJ. Compton. 2008. Comparing Mammalian Expression Systems: The First Rate-Limiting Step in Making Products for Clinical Testing. BioProcess Int. 6:S24-S32.
8.) Lindgren, K. 2009. Automation of Cell Line Development. Cytotechnol. 59:1-10.
9.) Andersson, P. 2007. Parallel Nanoliter Microfluidic Analysis System. Anal. Chem. 79:4022-4030.
You May Also Like





