Bacillus subtilis: A Promising Microbial Host System for Efficient Recombinant-Protein ProductionBacillus subtilis: A Promising Microbial Host System for Efficient Recombinant-Protein Production
September 1, 2024

High-throughput screening of alternative promoters, gene copy number, secretion-signal peptides, and protease-deficient strains can optimize product yields and stabilities, enabling g/L production of novel enzymes and therapeutic targets.
Recombinant-protein production is an important process for generating large quantities of biomolecules, including therapeutics, vaccines, and industrial enzymes (1). Mammalian cell expression systems — including Chinese hamster ovary (CHO) cells — are often the first choice of host system for biopharmaceutical applications. However, scaling up recombinant-protein production with mammalian cell lines can be time-consuming and costly because they require expensive media compositions, exhibit slow cell growth, and are highly sensitive to shear stresses (2). Using microbial hosts to express heterologous proteins allows for increased biomass densities at lower costs because such organisms require less expensive growth media. For example, eukaryotic organisms such as yeasts historically have been popular microbial hosts for recombinant-protein production because of their amenability to genetic manipulation and capacity to perform complex posttranslational modifications (PTMs), including glycosylation and the formation of disulfide bridges.
An Alternative Platform
Bacterial hosts such as Bacillus subtilis can offer even greater scale-up advantages compared with yeasts. Because of its faster growth rates and shorter fermentation cycles, using B. subtilis as a microbial host can lead to higher productivity. For instance, the organism can achieve a doubling time of as little as 20 minutes under an optimal growth temperature of 30–35 °C (3). Those conditions enable the typical B. subtilis fermentation cycle to be completed in ~48 hours, whereas the fermentation cycle of Saccharomyces cerevisiae is much longer at ~180 hours (1). B. subtilis occurs naturally in soil and has evolved to secrete a complex suite of enzymes that allow it to access nutrients in surrounding organic matter, compete with other organisms, and adapt to environmental changes (4, 5). Those abilities provide a convenient mechanism for recombinant-protein secretion into extracellular environments, which streamlines downstream processing for faster, more affordable protein purification.
In addition, the bacterium’s natural competence for DNA uptake and efficient homologous recombination makes it particularly suitable to genetic manipulation and highly adaptable to different manufacturer requirements. Ingenza has capitalized on those inherent capabilities by establishing a toolbox for genetically engineering B. subtilis. The technology includes a genome-editing platform using clustered regularly interspaced short palindromic repeats (CRISPR) and MAD7 enzymes (Inscripta), enabling the reprogramming of cellular metabolism to obtain desirable features and optimize expression (6).
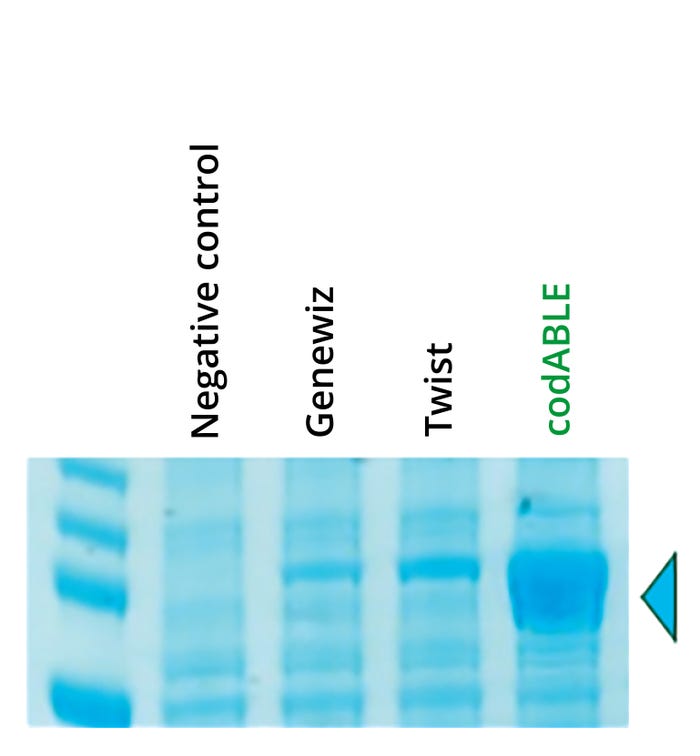
Figure 1: Sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDSPAGE) analysis of recombinant biocatalyst expression in Bacillus subtilis using alternative gene-design algorithms;
combining endotoxin-free production in B. subtilis with the codABLE technology’s optimized gene design provided superior.
Endotoxin-Free Protein Production System
Gram-negative hosts such as Escherichia coli produce endotoxins and other impurities that must be removed during downstream processing. Impurity removal requires complex purification steps that can increase overall production costs significantly (7–9). A major strength of B. subtilis is that it is a Gram-positive bacterium and thus does not produce endotoxins that can compromise biomedical applications. Consequently, naturally endotoxin-free protein production with B. subtilis is a cost-effective option and has enabled the organism to be granted generally regarded as safe (GRAS) status by the US Food and Drug Administration (FDA), as well as qualified presumption of safety (QPS) status by the European Food Safety Authority (EFSA) (4). Recognition from reputable organizations simplifies regulatory approvals for recombinant proteins produced by B. subtilis and enables their use for multiple human consumption and health-related applications. Table 1 presents a detailed, side-by-side comparison of E. coli and B. subtilis.
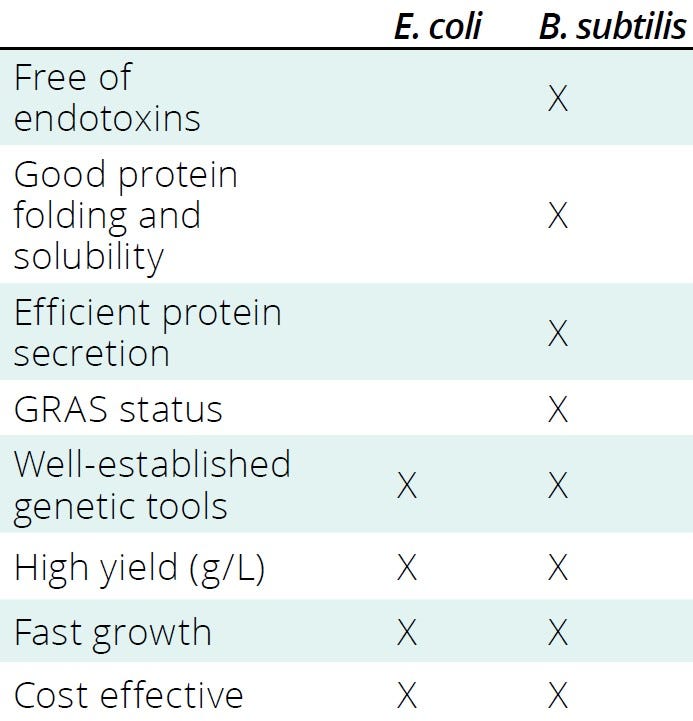
Table 1: Bacillus subtilis has several advantages over Escherichia coli as a microbial host system for recombinant protein expression; GRAS = generally regarded as safe.
Genetic Optimization for Enhanced Protein Production
Protein-expression levels are influenced by several factors, including the activity of promoters used to regulate gene transcription, the strength of the ribosome binding site, and the number of recombinant-gene copies. Therefore, those factors must be considered to ensure efficient gene expression (1). For example, each amino acid residue within a given protein can be encoded by multiple codons, and the choice of codon used has a strong effect on heterologous protein-expression levels. However, screening all possible gene variants for a protein of interest is unfeasible. Often, codon-optimization strategies are based on the relative abundance of each codon within the recipient manufacturing host, but evidence suggests that this is a simplistic and unreliable method (10). To address that challenge, Ingenza combined a large expression-data set with machine learning (ML) to develop the proprietary codABLE codon-optimization algorithm. The tool can predict optimal codon usage for expression of any target protein in B. subtilis and has been used successfully to improve the expression of multiple target molecules, consistently enhancing the expression of intracellular and secreted proteins.
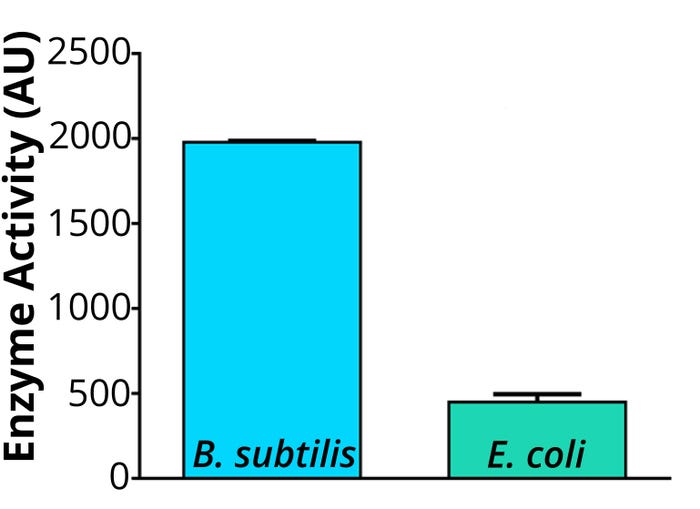
Figure 2: Enzyme-activity analysis in cell lysates of Ingenza’s Bacillus subtilis strain compared with an Escherichia coli strain; B. subtilis optimized production of a soluble biocatalyst candidate in the cell lysate.
The choice of a secretion signal peptide is also important for effective production in B. subtilis. Signal peptides can affect the translocation of recombinant proteins across cell membranes significantly, which in turn affects downstream purification. Secretion signal peptides are short amino acid sequences found at the N-terminus of newly synthesized secretory proteins and are specifically recognized by dedicated protein translocation machineries, playing a crucial role in directing proteins to extracellular media. Identifying optimal signal peptides for a particular protein sequence can be challenging. Certain combinations of proteins and signal peptides can lead to misfolding or improper processing of recombinant proteins in B. subtilis, affecting biological activity and stability. Ingenza therefore developed a library of secretion signal peptides as part of its engineering toolbox to overcome the difficulties of signal peptide selection. The library contains 94 diverse secretion signal peptides that can be fused to any gene sequence, allowing identification of optimal signal peptides to enhance protein translocation and secretion.
B. subtilis strains possess robust proteolytic systems, which pose a challenge when performing recombinant-protein expression. Some heterologous proteins are particularly susceptible to degradation during secretion. Ingenza has developed a collection of 21 protease-deficient strains — which together make up the Bacillus Ingenza Optimization (BINGO) platform — that can be screened for improved stability and production of challenging recombinant-protein targets in biopharmaceutical and industrial applications (4). Combining the codABLE and BINGO technologies with signal-peptide library screening enables Ingenza to double product yields and stabilities for g/L production of novel enzymes and therapeutic targets.

Bacillus subtilis offers many advantages for scale-up, including fast growth rates and short fermentation cycles.
A Diverse, Living Tool for Many Sectors
The versatility of B. subtilis has been demonstrated successfully by its use in expressing many protein products within pharmaceutical and medical fields — e.g., hyaluronic acid, antibodies and antibody fragments, surface growth factors, antibiotics, and lysozymes. Antimicrobial peptides expressed by B. subtilis also hold much promise with rapid, broad-spectrum killing activity against a number of pathogens, and the organism is used frequently to express heterologous antigens for vaccine manufacturing (1).
Another key application for B. subtilis is the industrial enzyme sector, which has an average annual turnover of >€2 billion in Europe, where the production of proteases for detergents alone accounts for 900 tons of pure enzymes per year (5). The substantial, growing demand for efficient enzymes with cost-effective manufacturing processes highlights a need for optimized secretion hosts. B. subtilis plays many roles in industrial applications because of its natural ability to secrete enzymes of commercial interest, including amylases, xylanases, and proteases. Amylases release simple sugars from starch — one of the most abundant, naturally occurring polymers — and are used widely in food, textiles, detergents, paper, and biofuel. Similarly, xylanases break down xylans to release oligosaccharide and disaccharide sugars and xylose, substances that are particularly valuable for modifying lignocellulosic materials in animal-feed manufacturing, biofuel production, and the paper and textile industries (1). Thanks to the bacterium’s GRAS and QPS statuses, protease enzymes produced by B. subtilis can be used in a number of food applications, including soybean and casein hydrolysate preparation, meat tenderization, milk coagulation, and even food-waste management (1).
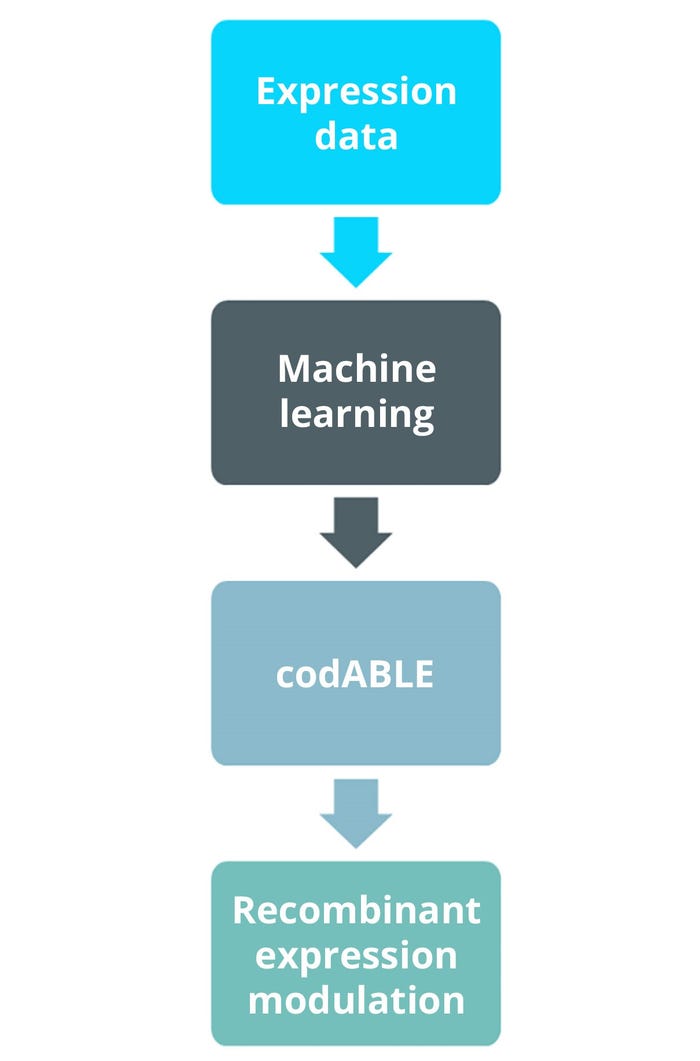
Figure 3: Workflow of Ingenza’s codABLE machine-learning (ML) developed algorithm.
Supporting the Transition to Green Manufacturing
A global increase in environmental consciousness is driving research into biological-based manufacturing processes, and microorganisms are being explored for their potential to enable more sustainable production methods than current hazardous and energy-intensive industrial approaches allow (4). The inherent secretory mechanisms of B. subtilis make it well suited to the production of various enzymes needed to establish green alternatives, including enzymes that enable the breakdown of abundant lignocellulosic biomass into simple, fermentable sugars. Those sugars then can be converted into biofuels, including ethanol and butanol, through fermentation.
Bacillus strains also have been engineered to produce D-lactic acid, a major component of renewable and biodegradable polylactic acid polymers with applications in packaging, textiles, and biomedical devices (11). That advancement points the way toward a more sustainable plastics industry with less dependence on conventional petrochemical-based plastics, which could help to remove some financial constraints associated with the manufacture of greener polymer-based goods (4). In addition, researchers are investigating the use of sustainable carbon sources — and even commercial waste materials — as feed sources for cultivating B. subtilis. Switching to renewable growth substrates and industrial by-products that would be discarded otherwise could contribute significantly to reducing the carbon footprint of manufacturing in many sectors, helping to establish a more circular global economy.
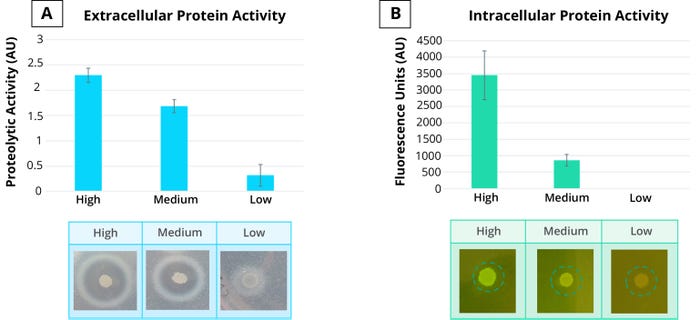
Figure 4: Ingenza’s codABLE gene-design algorithm enables modulation of recombinant-gene expression to control extracellular (left) and intracellular (right) protein expression.
Secreted protease activity on milk-protein–containing plates (left) and intracellular green fluorescent protein (GFP) (right) were used to confirm protein-expression predictions from the codABLE algorithm.
Opportunities for Future Exploration
The versatility of B. subtilis has made it the focus of intense research for decades. Many groups are seeking to discover novel approaches and applications, and one research avenue currently garnering a great deal of attention is the use of ML and artificial intelligence (AI) to identify how B. subtilis and other microbial hosts can be used to boost the expression of both proteins and small molecules. That could be particularly useful for products that require significant genetic and process optimization to overcome productivity, stability, and toxicity challenges. Overall, the organism offers many advantages relative to more established mammalian and microbial systems, with new uses continually being discovered and implemented. The full potential of B. subtilis as an alternative to E. coli is beginning to be realized, and the organism shows much promise for its contributions to human health and a more sustainable planet.

CUSTOMER CASE STUDY
Candidate protein
Enzyme used in pharmaceutical manufacturing
Challenges
Low yield from Escherichia coli as an insoluble aggregate
Host-cell endotoxin contamination
High cost of goods (CoG)
Complex purification
Solution
Alternate endotoxin-free Bacillus subtilis production host
Optimized gene design using codABLE algorithm
Results
Gene design with superior expression (see Figure 1)
Optimized production of candidate in soluble form (see Figure 2)
Cost-effective, scalable, endotoxin-free biomanufacturing process
Strain and process technology-transfer package

References
1 Su Y, et al. Bacillus subtilis: A Universal Cell Factory for Industry, Agriculture, Biomaterials and Medicine. Microb. Cell Fact. 19, 2020: 173; https://doi.org/10.1186/s12934-020-01436-8.
2 Rettenbacher LA, et al. Microbial Protein Cell Factories Fight Back? Trends Biotechnol. 40(5) 2022: 576–590; https://doi.org/10.1016/j.tibtech.2021.10.003.
3 Errington J, van der Aart LT. Microbe Profile: Bacillus subtilis: Model Organism for Cellular Development, and Industrial Workhorse. Microbiology 166(5) 2020: 425–427; https://doi.org/ 10.1099/mic.0.000922.
4 Yoshida K, van Dijl JM. Engineering Bacillus subtilis Cells as Factories: Enzyme Secretion and Value-Added Chemical Production. Biotechnol. Bioprocess Eng. 25(6) 2020: 872–885; https://doi.org/10.1007/s12257-020-0104-8.
5 van Dijl JM, Hecker M. Bacillus subtilis: From Soil Bacterium to Super-Secreting Cell Factory. Microb. Cell Fact. 12, 2013: 3; https://doi.org/10.1186/1475-2859-12-3.
6 Schneier M, et al. Current Technologies to Endotoxin Detection and Removal for Biopharmaceutical Purification. Biotechnol. Bioeng. 117(8) 2020: 2588–2609; https://doi.org/10.1002/bit.27362.
7 Mahmoodi S, et al. Current Affinity Approaches for Purification of Recombinant Proteins. Cogent Biol. 5(1) 2019: 1665406; https://doi.org/10.1080/23312025.2019.1665406.
8 Jozala AF, et al. Biopharmaceuticals from Microorganisms: From Production to Purification. Braz. J. Microbiol. 47, 2016: 51–63; https://doi.org/10.1016/j.bjm.2016.10.007.
9 Price MA, et al. Expanding and Understanding the CRISPR Toolbox for Bacillus subtilis with MAD7 and dMAD7. Biotechnol. Bioeng. 117(6) 2020: 1805–1816; https://doi.org/10.1002/bit.27312.
10 Tuller T, et al. An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell 141(2) 2010: 344–354; https://doi.org/10.1016/j.cell.2010.03.031.
11 Awasthi D, et al. Metabolic Engineering of Bacillus subtilis for Production of D-Lactic Acid. Biotechnol. Bioeng. 115(2) 2018: 453–463; https://doi.org/10.1002/bit.26472.
Rita Cruz, PhD, is section head for strain engineering at Ingenza Ltd., Bush Farm Road, Roslin EH25 9RG, Edinburgh, United Kingdom; [email protected].
codABLE is a registered trademark of Ingenza Ltd.
You May Also Like






