Safeguarding Bioproduction Modalities: Comprehensive Testing Strategies Require Reliable Analytical Processes and Reference MaterialsSafeguarding Bioproduction Modalities: Comprehensive Testing Strategies Require Reliable Analytical Processes and Reference Materials
Bioproduction technologies have revolutionized medicine, enabling the development of vaccines, cell and gene therapies (CGTs), and other biologics that hold promise for treating many diseases. Such modalities represent a dynamic, evolving pharmaceutical industry sector, and ensuring their safety and efficacy is crucial. Regulatory authorities and biopharmaceutical companies continuously seek to improve methods for detecting and identifying pathogens and other impurities in biotherapeutic products so that they can be safely inactivated or removed before administration to patients (1).
Integration of Testing Modalities in Bioproduction
Integrating multiple testing methods into bioproduction ensures comprehensive product-quality and safety evaluation. Rigorous testing protocols must be used to determine that the products are free of bacterial, fungal, mycoplasma, and viral contamination; bacterial endotoxins; and residual host cell DNA and proteins. Such tests serve as critical checkpoints throughout manufacturing processes, guaranteeing the quality and safety of therapeutic agents. Assessments are performed at different process stages, including raw-material testing, in-process monitoring, and final product release (Figure 1). Implementing robust analysis protocols helps to minimize the risk of product contamination, increases compliance with regulatory standards, and safeguards patient health.
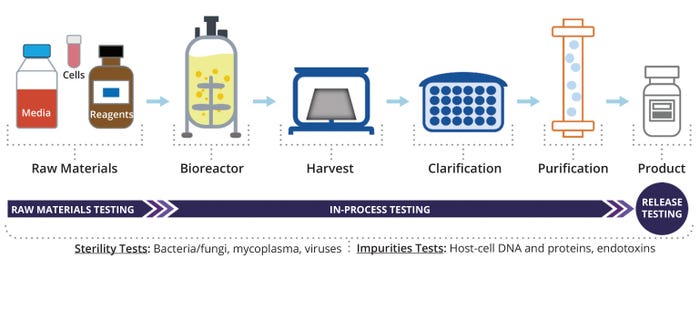
Figure 1: Workflow of impurity and contaminant testing during bioproduction.
Pharmacopoeias and regulatory agencies worldwide have collaborated on compendial harmonization for decades (2). The Pharmacopoeial Discussion Group (PDG) — formed in 1990 and comprising the European Pharmacopoeia (EP), Japanese Pharmacopoeia (JP), and United States Pharmacopeia (USP) — plays a key role in this activity. The PDG produces harmonized pharmacopoeial texts through independent public comment and consultation. In addition, the PDG reports on the progress of its harmonization efforts at the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) meetings. In November 2003, ICH established the Q4B Expert Working Group (EWG) to evaluate and recommend pharmacopoeial texts for use in the three ICH regions. The Q4B EWG anticipates that the PDG will be the principal source of pharmacopoeial text proposals. For example, when issuing Q6A and Q6B specifications, ICH recognized that the value and applicability of both guidelines would depend on the successful harmonization of pharmacopoeial procedures and encouraged the work of the PDG (3).
Classic and Rapid Microbiology Sterility Testing: Microbiology sterility testing is a critical aspect of bioproduction intended to detect the presence of undesirable microorganisms in pharmaceutical products. Classic sterility-testing methods, such as membrane filtration and direct inoculation, have been used in bioproduction for many years. Conventional bioburden and sterility testing methods are outlined in detail in USP Chapters <61>, <62>, and <71> (4–6); EP chapters 2.6.1, 2.6.12, and 2.6.13 (7–9); and JP chapters 4.05 and 4.06 (10–11). Such methods involve incubating samples under appropriate conditions that allow microbial growth. The positive controls recommended in those pharmacopoeial documents consist of known contaminants used to demonstrate that test media can support microbial growth (Table 1).
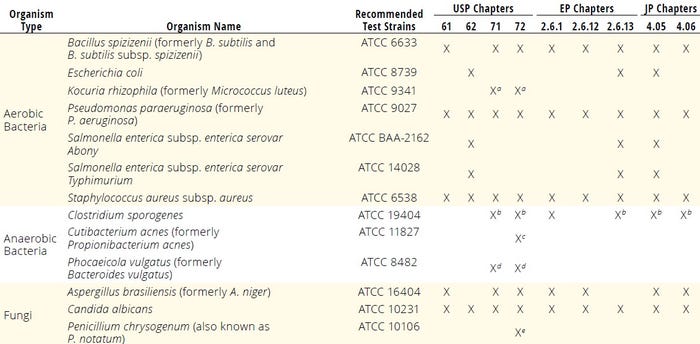
Table 1: Standard test organisms used for microbial enumeration, sterility, and rapid microbiological methods to confirm media growth promotion; EP = European Pharmacopoeia, JP = Japanese Pharmacopoeia, subsp. = subspecies, USP = United States Pharmacopeia.
a An alternative microorganism to Pseudomonas paraeruginosa b Clostridium sporogenes ATCC 11437 can also be used as an alternative. c Proposed
d An alternative microorganism to Clostridium sporogenes e Recommended when using a single incubation temperature
Although they are reliable, classic growth-based methods can be time consuming, requiring incubation periods of up to 14 days for typical microbial detection and even 28 days for mycoplasma assessments (6, 12). Innovative technologies such as respiration-based methods or nucleic-acid amplification techniques (NAATs) have emerged as promising alternatives to traditional approaches for rapid microbiology sterility testing (13). Such techniques offer significantly reduced turnaround times, enabling the much-desired timely release of products with short shelf lives (e.g., CGTs) while maintaining high levels of sensitivity and specificity. USP Chapter <72> outlines respiration-based rapid microbial methods for releasing short–shelf-life products and recommends positive control microorganisms for such tests (14).
EP Chapter 5.1.6 and USP Chapter <1223> (15, 16) guide the implementation and validation of alternative, rapid microbiological methods, ensuring both reliability and effectiveness. Critical considerations for ensuring the reliability of rapid microbiology sterility testing include method validation, equipment qualification, technology suitability, sample preparation of reliable biomaterials, and data analysis and interpretation. For a comprehensive review of all microbiological compendial methods, see Sandle (17).
Based on evaluation by the Q4B EWG, the ICH Steering Committee recommends that EP 2.6.12, JP 4.05, and USP <61> be used interchangeably in ICH regions (3). EP 2.6.1, JP 4.06, and USP <71> also can be used interchangeably in ICH regions subject to some conditions. For instance, diluting and rinsing fluids should not have antibacterial or antifungal properties if they are to be considered suitable for dissolving, diluting, or rinsing an article under sterility testing (3). Given the successful ICH Q4 achievements regarding classical microbiology tests, a natural direction of PDG efforts is now the harmonization of emerging microbial tests (e.g., rapid tests), including those for mycoplasmas.
Mycoplasma Testing and Cell-Culture Contamination Risks: Mycoplasmas are notorious bacterial contaminants of cell cultures and animal-derived products. Unlike other microbiological contaminants, mycoplasmas are not detected easily in cell cultures because they do not cause significant visible changes in cell-culture media (e.g., turbidity, pH, or cell death) (18). Mycoplasma contamination poses a significant concern in bioproduction because of its potential to compromise product safety and efficacy by affecting cell-culture performance and patient health (19).
Different pharmacopoeias provide detailed protocols for culture-based or NAAT mycoplasma testing to ensure product safety and efficacy (20–23). Table 2 lists recommended positive-control mycoplasmas described in those compendial documents.
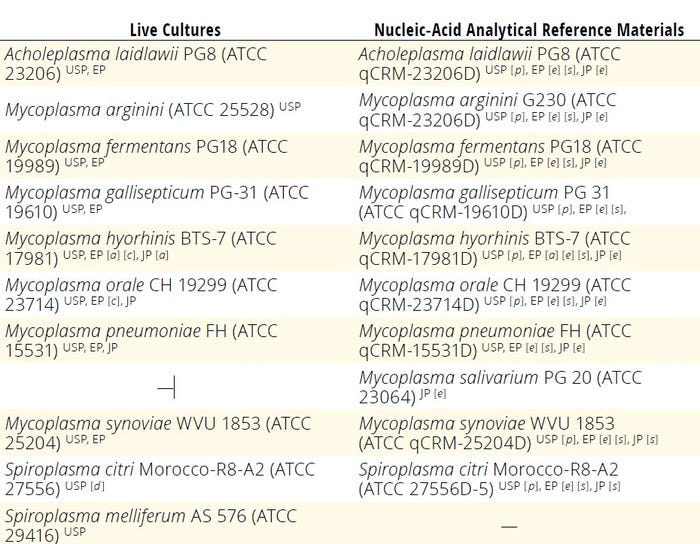
Table 2: Mycoplasma reference positive controls; EP = European Pharmacopoeia, JP = Japanese Pharmacopoeia, USP = United States Pharmacopeia.
[a] Mycoplasma hyorhinis ATCC 29052 can be used as an alternative.
[c] M. hyorhinis ATCC 29052 and M. orale ATCC 23714 are recommended for the cell-culture stain suitability method.
[d] Spiroplasma citri ATCC 29747 can be used as alternative.
[e] Nucleic-acid derivative from reference strain used for live culture or equivalent
[p] Proposed
[s] Stated as species without any particular biorepository reference
Testing for Adventitious Virus Contamination: Adventitious virus contamination poses a critical risk to biotherapeutic product quality and patient safety. Despite the industry’s safety efforts, viral infection of mammalian cell cultures remains a genuine risk that can lead to drug shortages and damaged public confidence (24–26). The concern has prompted manufacturers to implement virus-clearance methods such as filtration, chromatography, and inactivation with chemicals, detergents, or heat (26, 27). To demonstrate the efficacy of such methods, regulatory agencies have mandated that manufacturers perform rigorous testing using validated procedures to evaluate the successful removal or inactivation of viral contaminants (28–32). Another significant effort to address viral risk was the formation of the Consortium on Adventitious Agent Contamination in Biomanufacturing (CAACB) in 2011, which united over 20 pharmaceutical and biotechnology companies in that mission (33). The consortium aims to collect extensive data on viral contaminations of cell-culture operations from member companies and institutions. Over the past three decades, CAACB has identified 26 biochemical contaminations (24, 25).
Polymerase chain reaction (PCR) assays and high-throughput sequencing (HTS) commonly are used for adventitious virus detection in cell lines and biotherapeutic products (25, 34–36). Next-generation sequencing (NGS) has emerged as a potent detection tool. Unlike traditional NAAT assays targeting known viruses, HTS/NGS offers a comprehensive approach by sequencing a sample’s entire nucleic-acid content, thus capturing the virome in detail (36, 37). Remaining challenges in implementing NGS include data-analysis complexity, result interpretation, and the need for reliable reference materials for authentication (35). Table 3 summarizes adventitious viruses recommended as positive controls for cell-culture testing and virus-clearance validation studies (24, 28–32, 35, 37–39).
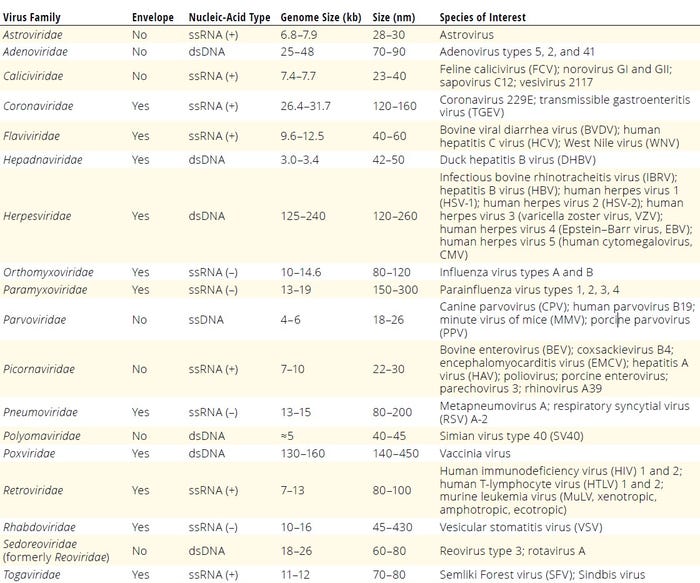
Table 3: Adventitious viruses recommended for cell-culture testing or virus-clearance validation studies; dsDNA = double-stranded DNA, ssRNA (+) = positive-stranded RNA, ssRNA (–) = negative-stranded RNA.
Endotoxin Testing and Product Safety: Endotoxins, primarily originating from the cell walls of Gram-negative bacteria, are a category of impurities can trigger severe immune reactions in patients. Therefore, stringent testing protocols and reliable methods for endotoxin detection are imperative to guarantee product safety and efficacy. The Limulus amebocyte lysate (LAL) assay, derived from horseshoe crab blood (Limulus polyphemus or Tachypleus tridentatus), stands as the primary method for endotoxin detection in pharmaceutical products (17, 40).
Control-standard endotoxins (CSEs) are endotoxin preparations traceable to international reference standards. They serve as secondary or tertiary standards certified by LAL reagent manufacturers for specific assay conditions. CSEs are used widely to prepare standard-curve calibrators and assay controls, offering cost savings and conserving primary standards. The US Food and Drug Administration (FDA) encourages use of suitably calibrated CSEs alongside international reference endotoxin standards (41). Based on the evaluation by the Q4B EWG, the ICH Steering Committee, recommends that the analytical procedures described in official pharmacopoeial texts — EP 2.6.14, JP 4.01, and USP General Chapter <85> — can be used interchangeably in ICH regions subject to some conditions. For instance, USP, JP, and EP reference standards are considered to be interchangeable because they have been suitably calibrated against the World Health Organization (WHO) international standard for endotoxins.
Authenticated Materials and Process Development
Using authenticated source materials and rigorous process-development methodologies is paramount in bioproduction. Authenticated cell lines, microorganisms, culture media, reagents, and other raw materials are the foundation for reproducible and reliable research, product development, and manufacturing processes (17, 18, 50). Certified reference standards and analytical reference materials can enable accurate quantification and ensure traceability to international standards.
Microbial Authentication: Biomaterial standards and reference materials are critical for sterility and impurity testing. Within microbiology laboratories, type strains or standards should be preserved carefully, and there should be no more than five transfers (passages) from the original culture to prevent phenotypic and genotypic divergences during long-term storage (4–12, 51). Pharmacopoeias recommend the use of microorganisms that are traceable to trustworthy repositories known for their reliability in biomaterial authentication processes — e.g., the American Type Culture Collection (ATCC), UK National Collection of Type Cultures (NCTC), German Collection of Microorganisms and Cell Cultures (DSMZ), Collection of the Institute Pasteur in France, UK National Collection of Industrial, Food and Marine Bacteria, Japanese NITE Biological Resource Center, UK National Collection of Pathogenic Fungi, and CABI (4–12, 17).
At ATCC, initial prokaryote characterization involves morphology and purity testing, biochemical analyses, protein-profile analyses using matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS), 16S ribosomal RNA (rRNA) or full-genome sequencing, and, in special cases, functional testing (e.g., antimicrobial resistance profiling) (52). For lot release, authentication criteria consist of viability, purity, and identity confirmed by MALDI-TOF MS and either 16S-rRNA or complete-genome sequencing. Initial characterization of fungi entails viability, purity, and morphology analyses as well as genotyping by sequencing of the internal transcribed spacers (ITS) ITS1 and ITS2, the 18S rRNA gene from the small subunit, and both the 5.8S rRNA gene and 5 ′end of the 28S rRNA gene (D1/D2 hyper variable region) from the large subunit depending on species. ITS and D1/D2 are commonly used (53). Batch release authentication for fungi and yeast entails viability assays, purity testing, and sequencing of the ITS or D1/D2 regions. For viruses, initial characterization consists of analyses for purity, viability, infectivity, certain virus-dependent characteristics (e.g., hemadsorption or hemagglutination), and sequencing (54). Lot-release quality control (QC) authentication for viruses entails purity testing, viability testing, and whole-genome sequencing. All sequencing results are compared with reference sequence genomes on the ATCC Genome Portal (55).
Cell-Line Authentication: Cell-line banking and characterization are the foundation of reliable bioproduction processes. Specific cell-line characterization and cell-banking chapters have been listed in several ICH guidelines since 1987. Such sources provide broad guidance on appropriate standards for preparing and characterizing human and animal cell lines to produce biological products. Guidelines emphasize the principle of the cell-bank system and the qualification tests of cell banks, which include cell authenticity and absence of bacteria, fungi, mycoplasmas, and viruses. Cell-bank preparation operations should comply with good manufacturing practice (GMP). It must undergo rigorous characterization testing to ensure the purity, safety, functionality, and genetic stability of cells grown from those banks (56). Cell authentication is critical to cell-line development, cell banking and downstream bioprocessing. Once a new cell line is developed, cell banks are propagated and used to produce biologicals. It is important to perform cell-line authentication from beginning to end to make sure that master and working banks remain the same so that bioproduction starts with the correct cells every time.
Several mammalian cell lines — e.g., human embryonic kidney (HEK) 293, Vero, Madin–Darby canine kidney (MDCK), and Chinese hamster ovary (CHO) K1 — are used extensively as host cell lines for biological product manufacturing. Because the safety and quality of biologics developed for therapeutic use are paramount, host cell lines used in biomanufacturing need to be authenticated to ensure their identity is verified and that they are free of contamination by other cell lines, viruses, and microbes. Authentication of a cell line refers to the sum of processes by which it is confirmed to be from the correct species and donor. Rough estimates suggest that the number of published studies involving problematic cell lines is about 16.1% (18, 50, 57). Moreover, the International Cell Line Authentication Committee (ICLAC) lists 576 misidentified or cross-contaminated cell lines in its latest register (18). Cell-line authentication is one of the most critical steps for high-quality cell culture results. Whether as a result of initial cell-line misidentification, cross-contamination, or genetic drift, the result is the same: wasted money and resources.
Interspecies Identification: Inter- and intraspecies identification tests can verify a cell line’s identity, including its species and original donor. Such methods can determine the species of a tested cell line and whether it is mixed with cells from other species. To achieve species identification, an ideal marker should have attributes such as being highly conserved among species, displaying enough variation to allow species-level discrimination. Practitioners should use markers that are listed in a database containing verified DNA sequences. Cytochrome c oxidase subunit 1 (CO1) DNA sequencing and multiplex PCR assays are two methods designed for interspecies identification. CO1 has met all requirements, and the Consortium for the Barcode of Life has adopted it for standard DNA metabarcoding. CO1 is a mitochondrial gene expressed in all animal species. Because the inheritance of mitochondrial genes is maternal, animals typically have only one variant of each mitochondrial gene. Through CO1 DNA sequencing, a cell line’s species can be determined precisely. In addition, because cells from different species have different sizes of CO1, a multiplex PCR assay can determine the species of a cell culture easily. Users can screen for species cross-contamination events by simply running multiplex PCR amplification products on a DNA gel. For instance, ATCC scientists developed a PCR-based speciation assay by designing unique primer pairs that are species-specific and produce amplicons in a multiplex PCR reaction ≥20 base pairs apart. The ATCC CO1 assay can distinguish cell lines of several mammalian origins (58).
Intraspecies Donor-Level Identification: Intraspecies identification separates cells originating from different donors, even if they are from the same species. Short-tandem repeat (STR) profiling of cell lines is the gold standard for both intraspecies donor-level identification and identity confirmation for human cell lines.
STR profiling initially was developed for identity tests in the forensics field. The unique STR profile of each human cell line is based on the informativeness and variation of tandem repetitive sequences in the genomes of different cell lines. Such profiles serve as essential components in conducting valid, reproducible, and meaningful research. STR DNA is microsatellite DNA, and the elements are regions of repeated DNA sequences that are found throughout eukaryotic genomes. The number of repeats in STR loci vary greatly among individuals, making such genetic markers effective for human identification or cell line authentication. STR markers are used for DNA identity testing because they are highly polymorphic and easily amplified by PCR, and electrophoresis can distinguish amplicons by their size differences. When multiple STR markers are examined simultaneously, a DNA profile can be created to allow unambiguous sample identification. The STR assay is highly sensitive because the DNA amplicons are tagged fluorescently, and the amplicons’ detection and size calculations by capillary electrophoresis are highly accurate.
In addition to being fast and reliable, STR tests are robust. For each STR marker added to the pool, the power of discrimination improves by about an order of magnitude. Multiple STR loci (typically 8–16) loci and amelogenin (used for sex determination) are amplified and compared against a reference database to ensure accurate identification. The discrimination power of 16 loci STR profiling is approximately 10–22, which means the probability of a random match using 16 STR markers between two cell lines from different individuals is approximately 10–22.
Furthermore, STR DNA fragment sizes can be converted to numerical values corresponding to the number of repeats in each allele. Thus, the STR profile of a given cell line can be compared among different laboratories easily. To eliminate the risk of misidentification and cross-contamination, STR testing of human cell lines is conducted routinely by many cell repositories, international cell banks, and biological resource centers such as ATCC, DSMZ, and the European Collection of Authenticated Cell Cultures (ECACC). As a result, the STR profiles of thousands of human cell lines have been collected and made publicly available. A comprehensive and definitive consensus standard regarding STR profiling — ASN-0002-2022 — also has been published. It includes best practices for STR profiling, data analyses and QC, interpretation of the results, and implementation of a searchable public database (59, 60).
Although STR profiling is extremely powerful for authentication of human cell lines, it has its own limitations. It cannot discriminate among cells derived from monozygotic identical twins, cells derived from different tissues of the same donor, or engineered cells from the same parental cell line. Additional genotyping assays are required for authenticating genetically modified and isogenic cell lines. Some host cell lines used in bioprocesses have been modified for improved bioproduction capabilities, either through a simple cell-cloning approach or more complex genetic modification. Traceability from the original parent-cell lines to the established new derivative lines is essential for manufacturing final biological products and establishing host cell-line genomic DNA (gDNA) and host cell protein (HCP) reference materials.
It is worth noting that, in addition to cell characterization, purity and safety have been emphasized with the encouragement of new technology in recent FDA guidelines. In the second revision of the ICH Q5A guidance regarding viral safety of biotechnological products derived from characterized cell lines of human or animal origin (28, 29), the EWG provided more details. Those included considerations for alternative analytical methods as well as clearer recommendations on when and how to test master cell banks (MCBs), working cell banks (WCBs), seed stocks, cells at the limit of in vitro cell age (LIVCAs), and bulk harvest for adventitious viruses. Global regulatory bodies encourage biopharmaceutical developers to use new test methods based on NGS and NAATs to reduce animal use in drug testing — the 3R principle of replacement, reduction, and refinement. (61, 62). At ATCC, lot-release testing for cell lines and their derivatives (e.g., gDNA) includes authentication in addition to purity testing (e.g., absence of contaminants such as viruses, mycoplasma, bacteria, and fungi) (32, 60).
Crucial for Quality Control
Advancements in bioproduction technologies have revolutionized medicine, offering innovative treatments for many diseases. However, such new therapies require changes in bioproduction practices as well as robust, rapid testing to meet stringent regulatory standards for safety and efficacy. The latter factor is particularly crucial in fast-paced CGT protocols, which often involve products with short shelf lives, requiring microbiological QC tests to ensure timely, safe, and effective treatments.
QC testing is pivotal in establishing reliable development and full-scale manufacturing processes for safe and effective biologics. Developing and sustaining a robust QC pipeline for bioproduction modalities require access to reliable standards and analytical reference materials. Such materials include authenticated live microorganisms and cell lines, highly purified viruses, host cell DNA and proteins, and endotoxins. By integrating and harmonizing validated testing protocols with authenticated materials and standards, biomanufacturers can uphold the highest level of product quality, regulatory compliance, and patient welfare.
References
1 Ill CR, Dehghani H. Risk Reduction in Biotherapeutic Products. Curr. Opin. Drug Discov. Devel. 12(2) 2009: 296–304.
2 Wiggins JM, Albanese JA. Harmonization Efforts by Pharmacopoeias and Regulatory Agencies. BioPharm Int. 15 September 2019; https://www.biopharminternational.com/view/harmonization-efforts-pharmacopoeias-and-regulatory-agencies-2.
3 ICH Q4B Annex 4B. Evaluation and Recommendation of Pharmacopoeial Texts for Use in the ICH Regions on Tests for Specified Micro-Organisms. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use: Brussels, Belgium, 2010; https://database.ich.org/sites/default/files/Q4B%20Annex4B%28R1%29%20Guideline.pdf.
4 USP Chapter <61>. Microbiological Examination of Nonsterile Products: Microbial Enumeration Tests. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.org/10.31003/USPNF_M98800_01_01.
5 USP Chapter <62>. Microbiological Examination of Nonsterile Products: Test for Specified Organisms. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.org/10.31003/USPNF_M98802_01_01.
6 USP Chapter <71>. Sterility Tests. United States Pharmacopoeial Convention: Rockville, MD, 2024; https://doi.org/10.31003/USPNF_M98810_01_01.
7 EP 2.6.1. Sterility. European Pharmacopoeia: Strasbourg, France, 2020.
8 EP 2.6.12. Microbiological Examination of Non-Sterile Products: Microbial Enumeration. European Pharmacopoeia: Strasbourg, France, 2020.
9 EP 2.6.13. Microbiological Examination of Non-Sterile Products: Test for Specified Microorganisms. European Pharmacopoeia: Strasbourg, France, 2020.
10 JP 4.05. Microbiological Examination of Non-Sterile Products. 17th Ed. Pharmaceuticals and Medical Devices Agency: Tokyo, Japan, 1 April 2016; https://www.pmda.go.jp/files/000217650.pdf.
11 JP 4.06. Sterility Test. 17th Ed. Pharmaceuticals and Medical Devices Agency: Tokyo, Japan, 1 April 2016; https://www.pmda.go.jp/files/000217650.pdf.
12 ORA.007. Pharmaceutical Microbiology Manual. US Food and Drug Administration: Rockville, MD, 2020; https://www.fda.gov/media/88801/download.
13 Lin NJ, et al. Report from the 2022 NIST Rapid Microbial Testing Methods (RMTM) Workshop. National Institute of Standards and Technology: Gaithersburg, MD, 2023; https://doi.org/10.6028/NIST.SP.1292.
14 USP Chapter <72>. Respiration-Based Rapid Microbial Methods for the Release of Short Shelf-Life Products. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.usp.org/USPNF/USPNF_M16055_20101_01.html
15 EP 5.1.6. Alternative Methods for Control of Microbiological Quality. European Pharmacopoeia: Strasbourg, France, 2020.
16 USP Chapter <1223>. Validation of Alternative Microbiological Methods. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.org/10.31003/USPNF_M99943_04_01.
17 Sandle T. Pharmaceutical Microbiology: Essentials for Quality Assurance and Quality Control. Woodhead Publishing: Sawston, UK, 2015.
18 Weiskirchen S, et al. A Beginner’s Guide to Cell Culture: Practical Advice for Preventing Needless Problems. Cells 12(5) 2023: 682; https://doi.org/10.3390/cells12050682.
19 Smith CC, Demick JM. Mycoplasma Testing in an Ever-Changing World. Am. Pharm. Rev. 24 March 2022; https://www.americanpharmaceuticalreview.com/Featured-Articles/584555-Mycoplasma-Testing-in-an-Ever-Changing-World/.
20 USP Chapter <63>. Mycoplasma Tests. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.usp.org/USPNF/USPNF_M3687_01_01.html.
21 USP Chapter <77>. Mycoplasma Nucleic Amplification Tests. United States Pharmacopeial Convention: Rockville, MD, 2024.
22 EP 2.6.7. Mycoplasmas. European Pharmacopoeia: Strasbourg, France, 2020.
23 JP G3. Mycoplasma Testing for Cell Substrates Used for the Production of Biotechnological/Biological Products. 17th Ed. Pharmaceuticals and Medical Devices Agency: Tokyo, Japan, 1 April 2016; https://www.pmda.go.jp/files/000217650.pdf.
24 Barone PW, et al. Viral Contamination in Biologic Manufacture and Implications for Emerging Therapies. Nat. Biotechnol. 38(5) 2020: 563–572; https://doi.org/10.1038/s41587-020-0507-2.
25 Riske F. Microorganism Contamination in Downstream Biopharmaceutical Processes: A Holistic Approach to Measurement, Control, and Prevention. BioProcess Int. 22(3) 2024: 35–41; https://www.bioprocessintl.com/separation-purification/microorganism-contamination-in-downstream-biopharmaceutical-processes-a-holistic-approach-to-measurement-control-and-prevention.
26 Liu S, et al. Development and Qualification of a Novel Virus Removal Filter for Cell Culture Applications. Biotechnol. Prog. 16(3) 2000: 425–434; https://doi.org/10.1021/bp000027m.
27 Ray S, Tarrach K. Virus Clearance Strategy Using a Three-Tier Orthogonal Technology Platform. BioPharm Int. 22(6) 2009; https://www.biopharminternational.com/view/virus-clearance-strategy-using-three-tier-orthogonal-technology-platform.
28 Q5A (R2). Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use: Brussels, Begium, 2023; https://database.ich.org/sites/default/files/Q5A_R1_Guideline.pdf
29 Q5A (R2). Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. US Food and Drug Administration: Rockville, MD, 2024; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/q5ar2-viral-safety-evaluation-biotechnology-products-derived-cell-lines-human-or-animal-origin.
30 Points To Consider in the Manufacture and Testing of Monoclonal Antibody Products for Humans. US Food and Drug Administration: Rockville, MD, 1997; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/points-consider-manufacture-and-testing-monoclonalantibody-products-human-use.
31 Content and Review of Chemistry, Manufacturing, and Control (CMC) Information for Human Somatic Cell Therapy Investigational New Drug Applications (INDs). US Food and Drug Administration: Rockville, MD, 2008; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/content-and-review-chemistry-manufacturing-and-control-cmc-information-human-somatic-cell-therapy.
32 Guidance for Industry: Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications. US Food and Drug Administration: Rockville, MD, 2010; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/characterization-and-qualification-cell-substrates-and-other-biological-materials-used-production.
33 A United Front. Nature 472(7344) 2011: 389–390; https://doi.org/10.1038/472389b.
34 Lovatt A. Applications of Quantitative PCR in the Biosafety and Genetic Stability Assessment of Biotechnology Products. J. Biotechnol. 82(3) 2002: 279–300; https://doi.org/10.1016/s1389-0352(01)00043-5.
35 Mee ET, et al. Development of a Candidate Reference Material for Adventitious Virus Detection in Vaccine and Biologicals Manufacturing by Deep Sequencing. Vaccine 34(17) 2016: 2035–2043; https://doi.org/10.1016/j.vaccine.2015.12.020.
36 Barone PW, et al. Historical Evaluation of the In Vivo Adventitious Virus Test and Its Potential for Replacement with Next Generation Sequencing (NGS). Biologicals 81, 2023: 101661; https://doi.org/10.1016/j.biologicals.2022.11.003.
37 Khan AS, et al. Report of the Third Conference on Next-Generation Sequencing for Adventitious Virus Detection in Biologics for Humans and Animals. Biologicals 83, 2023: 101696; https://doi.org/10.1016/j.biologicals.2023.101696.
38 Gombold J, et al. Systematic Evaluation of In Vitro and In Vivo Adventitious Virus Assays for the Detection of Viral Contamination of Cell Banks and Biological Products. Vaccine 32(24) 2014: 2916–2926; https://doi.org/10.1016/j.vaccine.2014.02.021.
39 Asher D, et al. Technical Report No. 47, Preparation of Virus Spikes Used for Virus Clearance Studies. Parental Drug Association: Bethesda, MD, 2010.
40 USP Chapter <85>. Bacterial Endotoxins Test. United States Pharmacopeial Convention: Rockville, MD, 2024.
41 Guidance for Industry: Pyrogen and Endotoxins Testing: Questions and Answers. US Food and Drug Administration: Rockville, MD, 2012; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/guidance-industry-pyrogen-and-endotoxins-testing-questions-and-answers.
42 Evaluation of Medicinal Products for Human Use (EMEA) Position Statement on the Use of Tumourigenic Cells of Human Origin for the Production of Biological and Biotechnological Medicinal Products. The European Agency for the Evaluation of Medicinal Products. CPMP/BWP/1143/00, 27/02/2001.
43 Zhang W, et al. Development and Qualification of a High Sensitivity, High Throughput Q-PCR Assay for Quantitation of Residual Host Cell DNA in Purification Process Intermediate and Drug Substance Samples. J. Pharm. Biomed. Anal. 100, 2014: 145–149; https://doi.org/10.1016/j.jpba.2014.07.037.
44 Wang Y, et al. Quantification of Residual BHK DNA by a Novel Droplet Digital PCR Technology. J. Pharm. Biomed. Anal. 159, 2018: 477–482; https://doi.org/10.1016/j.jpba.2018.07.022.
45 USP Chapter <509>. Residual DNA Testing. United States Pharmacopeial Convention: Rockville, MD, 2024.
46 USP Chapter <1132>. Residual Host Cell Protein Measurement in Biopharmaceuticals. United States Pharmacopeial Convention: Rockville, MD, 2024; https://doi.org/10.31003/USPNF_M8647_01_01.
47 Bracewell DG, Francis R, Smales CM. The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control. Biotechnol. Bioeng. 112(9) 2015: 1727–1737. https://doi.org/10.1002/bit.25628.
48 Zhu-Shimoni J, et al. Host Cell Protein Testing by ELISAs and the Use of Orthogonal Methods. Biotechnol. Bioeng. 111(12) 2014: 2367–2379. https://doi.org/10.1002/bit.25327.
49 Tscheliessnig AL, et al. Host Cell Protein Analysis in Therapeutic Protein Bioprocessing — Methods and Applications. Biotechnol. J. 8(6) 2013: 655–670. https://doi.org/10.1002/biot.201200018.
50 Souren NY, et al. Cell Line Authentication: A Necessity for Reproducible Biomedical Research. EMBO J. 41(14) 2022: e111307; https://doi.org/10.15252/embj.2022111307.
51 Snell JJS. Preservation of Control Strains. Quality Assurance: Principles and Practice in the Microbiology Laboratory. Snell JJS, Brown DFB, Roberts C, Eds. Public Health Laboratory Service: London, UK, 1995: 69–76.
52 Authentication of Prokaryotes at ATCC. ATCC: Washington, DC, 2024; https://www.atcc.org/resources/white-papers/authentication-of-prokaryotes-at-atcc.
53 ATCC Mycology Culture Guide. ATCC: Washington, DC, 2024; https://www.atcc.org/resources/culture-guides/mycology-culture-guide.
54 ATCC Virology Culture Guide. ATCC: Washington, DC, 2024; https://www.atcc.org/resources/culture-guides/virology-culture-guide.
55 Benton B, et al. The ATCC Genome Portal: Microbial Genome Reference Standards with Data Provenance. Microbiol. Resour. Announc. 10, 2021: e00818-21; https://doi.org/10.1128/MRA.00818-21.
56 Gazaille B, Castillo F, Nims RW. Cell Banking in the Spotlight: Advising Biologics Developers About Cell Bank Preparation and Characterization. BioProcess Int. 19(6)i 2021: 1–4; https://www.bioprocessintl.com/cell-line-development/cell-banking-in-the-spotlight-advising-biologics-developers-about-cell-bank-preparation-and-characterization
57 Babic Z, et al. Incidences of Problematic Cell Lines Are Lower in Papers That Use RRIDs To Identify Cell Lines. Elife 8, 2019: e41676; https://doi.org/10.7554/eLife.41676.
58 Cooper JK, et al. Species Identification in Cell Culture: A Two-Pronged Molecular Approach. In Vitro Cell Dev. Biol. Anim. 43(10) 2007: 344–351. https://doi.org/10.1007/s11626-007-9060-2.
59 Barrett T, et al. ANSI/ATCC ASN-0002-2022: Authentication of Human Cell Lines: Standardization of STR Profiling.
60 ATCC Animal Cell Culture Guide. ATCC: Washington, DC, 2024; https://www.atcc.org/resources/culture-guides/animal-cell-culture-guide.
61 Perez-Caballero D, et al. Viral Safet for Biotechnology Products, Including Viral Vectors: ICH Q5A Revision 2 Brings Updated and More Comprehensive Guidance. BioProcess Int. 21(5) 2023: 32–36; https://www.bioprocessintl.com/viral-clearance/viral-safety-for-biotechnology-products-including-viral-vectors-ich-q5a-revision-2-brings-updated-and-more-comprehensive-guidance.
62 EMA/CHMP/CVMP/JEG-3Rs/450091/2012. Guideline on the Principles of Regulatory Acceptance of 3Rs (Replacement, Reduction, Refinement) Testing Approaches. European Medicines Agency: Amsterdam, the Netherlands, 2012; https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-principles-regulatory-acceptance-3rs-replacement-reduction-refinement-testing-approaches_en.pdf.
Leka Papazisi, DVM, PhD, is principal scientist for research and industrial solutions, and Fang Tian, PhD, is director of biological content, both at ATCC; 10801 University Boulevard, Manassas, VA 20110.
You May Also Like







