Antibody and Protein Therapeutic Kinetic Characterization in BioprocessingAntibody and Protein Therapeutic Kinetic Characterization in Bioprocessing
July 1, 2008

Figure 1.
There are a variety of affinity tag-protein strategies, and the use of affinity tags depends on the target protein, the expression system, and the application. The Octet QK and Octet RED systems provide an easy-to-use, label-free, high-throughput system to capture affinity-tagged biologics for quantitative or kinetic biomolecular interaction analysis. Kinetic characterization of biologics during clone selection, screening, and development is critical at all stages of therapeutic engineering and bioprocessing.
Key Applications
Quantitative kinetic characterization (kobs′ ka′ kd′ KD — without flow-cell fluidics)
Binding affinity determination
High-throughput for rank ordering of clones (kinetic screening of 96 samples in two hours)
Epitope mapping
Binding pair selection for protein specific assay development
Principle
Protein binding and dissociation events can be monitored by measuring the binding of one protein in solution to a second protein immobilized on the FortéBio biosensor.
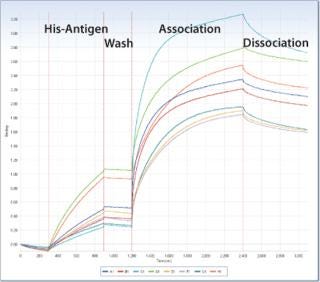
Figure 1. ()
Key Features
Key features of the Octet QK and Octet RED systems for kinetic screening and characterization include the following:
Label-free, real-time analysis: Using BLI technology, protein binding is continuously measured at the biosensor surface throughout the interaction and is displayed in real time.
Increased throughput: With parallel processing in a 96-well format, higher sample throughput for kinetics is easily achieved.
Simple assay set-up: With this fluidics-free method, assay development time is shortened with minimal instrument maintenance.
Flexible assay design: Customize workflow needs with either online immobilization for full kinetic characterization or offline batch immobilization for kinetic screening.
Direct readings from crude sample media: Eliminate time spent on sample purification or preparation.
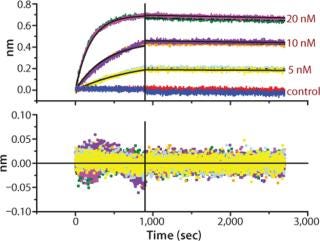
Figure 2. ()
You May Also Like






