A Platform for Purifying Pharmaceutical-Grade Plasmid DNA Using CIM® Monolithic ColumnsA Platform for Purifying Pharmaceutical-Grade Plasmid DNA Using CIM® Monolithic Columns

Figure 1.
CIM Convective interaction Media® are monolithic chromatographic supports used for purification of plasmid DNA, viruses, and other large biomolecules on a laboratory and industrial scale. CIM® monoliths are characterized by large flow-through channels (average diameter of 1,500 nm), which enhance the mass transfer between the mobile and stationary phase. This results in a high and flow-independent dynamic binding capacity and resolution. For plasmid DNA purification on an industrial level, these characteristics increase productivity, making CIM® monolithic columns attractive for industrial applications (1).
BIA Separations has developed a CIM®-based pharmaceutical-grade plasmid DNA purification process that can be used on both laboratory and industrial scales. One key step was the development of a new hydrophobic-interaction monolith, CIM® C4 (butyl). This HIC step results in the removal of open circular plasmid DNA from the supercoiled form, thus producing a final plasmid DNA product with >98% of the supercoiled form. Here we present the purification of 40 mg of plasmid DNA on a CIM® DEAE-8 (8 mL) followed by a CIM® C4-8 (8 mL) monolithic column.
Sample Preparation
The plasmid DNA purification process (Figure 1) starts with cultivation of Escherichia coli transformed with a defined plasmid. Harvested bacterial cells are lysed in alkaline conditions releasing the plasmid DNA. The lysate is treated with a neutralizing solution to precipitate host cell proteins and genomic DNA. The sample is further adjusted to 0.5 M CaCl2 for the removal of the majority of RNA, proteins, gDNA, and endotoxins. Clarification of the lysate is performed by centrifugation and/or filtration. Before sample loading onto the CIM® DEAE column, the conductivity is decreased to 37 mS/cm by diluting with sterile, deionized water.
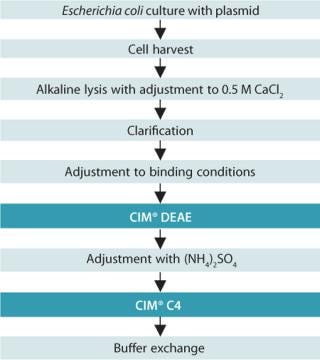
Figure 1. ()
Chromatography
To purify 40 mg of plasmid DNA, one CIM® DEAE-8 and one CIM® C4-8 monolithic column are used. In the first step, the CIM® DEAE column is equilibrated with 10 CV of the binding buffer at a flow rate of 125 mL/min (16 CV/min), and the clarified cell lysate is then loaded at the same flow rate onto the column. The remaining RNA and proteins bound to the column are washed with 0.6 M NaCl. The plasmid DNA is eluted using a stepwise gradient of 1.0 M NaCl (Figure 2). The eluted plasmid DNA is appropriate for laboratory analyses as >90% of the major impurities (genomic DNA, RNA, host cell proteins, and endotoxins) are removed (Table 1).
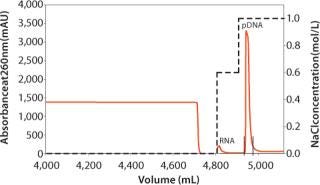
Figure 2. ()
Table 1. Analysis of fractions from the CIM® based plasmid DNA purification process
To increase the purity and obtain a higher percentage of supercoiled plasmid DNA (sc pDNA), a second purification step is required to target the removal of open circular plasmid DNA (oc pDNA). The plasmid DNA fraction eluted from the CIM® DEAE-8 column is adjusted to 3M (NH4)2SO4 and loaded onto an equilibrated CIM® C4-8 column at 125 ml/min (16 CV/min). After loading the OC pDNA is washed out using 1.7M (NH4)2SO4 and sc pDNA is eluted with 0.4M (NH4)2SO4.
Finally, the column is washed with regeneration buffer to elute any remaining genomic DNA (Figure 3). Because the dynamic binding capacity of the CIM® DEAE-8 column is higher than that of the CIM® C4-8 column, the HIC step needs to be performed twice to process all plasmid DNA that was purified in the first chromatographic step. Fractions containing sc pDNA are than combined for a buffer-exchange step. At laboratory scale, pDNA can be precipitated with isopropanol, but at pilot and production scales size-exclusion chromatography or tangential-flow filtration are preferred.
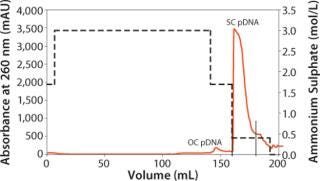
Figure 3. ()
Results
The resulting plasmid DNA purification process based on two CIM® monolithic columns yields high-quality product with >99% of all major contaminants removed: RNA, genomic DNA, host cell proteins, endotoxins, and open circular plasmid DNA (Table 1, Figure 4). This process can be completed in a matter of hours and is easily scalable. These factors in combination with high dynamic binding capacity and low buffer consumption significantly improve the economics of the process.
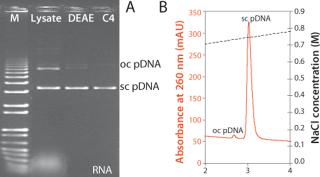
Figure 4. ()
REFERENCES
1.) Urthaler, J. 2005. Application of Monoliths for Plasmid DNA Purification: Development and Transfer to Production. J. Chromatog. A 1065:93-106.
You May Also Like






