Fc-Fusion Protein Capture Purification: Comparing an Innovative Membrane Adsorber Against Standard Column and Resin MaterialsFc-Fusion Protein Capture Purification: Comparing an Innovative Membrane Adsorber Against Standard Column and Resin Materials

Small-scale membrane chromatography device used in this study.
Antibodies play a crucial role in the human immune system, giving their therapeutic use and utility enormous potential in many respects (1). Thus, the production of antibodies and their derivatives — e.g., fusion proteins based on the fragment crystallizable (Fc) region — has been the focus of the biopharmaceutical industry for decades. Development of purification technologies for monoclonal antibodies (mAbs) made them feasible as a therapeutic product class (2, 3). As product concentrations in the one- to two-digit g/L scale have become state of the art, the upstream mAb production process has become less limited over time (4). By contrast, downstream processing must be optimized to handle such high expression titers and to reduce associated costs (5–7).
Today’s standard mAb purification processes consist of at least two chromatographic steps, with protein A affinity chromatography often used as an initial capture step because of its high affinity and selectivity for the Fc region of antibodies (8). Traditionally, packed columns of porous beads have been used to separate target proteins (9). However, certain disadvantages come with using bead-based chromatography columns, especially as production scale and titers increase. Concerns include limitations in productivity due to slow, diffusive mass transport or restrictions in scalability, as well as high cleaning and storage costs (9, 10).
Many such challenges can be overcome with membrane chromatography. One of the most decided advantages of membrane adsorbers — e.g., those with surfaces that are conjugated with protein A ligands — is their enablement of convection-based mass transport (thus speeding purification) through the altered geometry and expanded pore size of their material. However, a material based purely on convection can lose binding capacity. The Sartobind Rapid A membrane adsorber used in our experiments is based on a “convecdiff” material that combines the advantages of both convective- and diffusive-based mass transport (11).
Membrane adsorbers do not need to be packed, coming as ready-to-use devices that save related time and costs. Moreover, when used as single-use products, membrane adsorbers can increase safety and sustainability of downstream processing by eliminating cross-contamination, cleaning, and associated validation work (6, 7, 9, 10). Repetition of many short cycles can compensate for the lower matrix volume (12).
Here we investigate how the aforementioned advantages of membrane chromatography based on protein A can be implemented practically. For that purpose, the following study compares a capture process for a low-titer Fc-fusion protein on Sartobind Rapid A membrane adsorbers with a benchmark resin-based process.
Materials and Methods
Capture Development at Small (Laboratory) Scale: For small-scale capture development, we used a Sartobind Rapid A nano prototype (4-mm bed height and 1-mL membrane volume) provided by Sartorius Stedim Biotech. The membrane adsorber was connected to an ÄKTA avant 25 chromatography system controlled by Unicorn 6.1 software, both from Cytiva. The membrane adsorber and the chromatography system were prepared and stored as recommended by their suppliers.
Before loading, cell-culture supernatant was thawed at room temperature and sterile-filtered using Sartobran 300 filters (Sartorius Stedim Biotech) with a 0.2-µm cutoff. The supernatant was cooled throughout the entire capture period. Buffers were prepared in-house by dissolving chemicals from VWR International, Carl Roth GmbH + Co. KG, AppliChem GmbH, Merck Chemicals GmbH, and Fisher Scientific in deionized water from Bielefeld University. After some optimization work, equilibration, washing, and reequilibration were performed using phosphate-buffered saline (PBS). Elution steps 1 and 2 used citrate buffer, and clean-in-place (CIP) operations used sodium hydroxide (NaOH). Eluted product was neutralized with phosphate buffer.
The duration and volumes of each capture step were optimized in a handful of experiments, leading to the final recipe in Table 1. Following preliminary tests for characterization of the membrane adsorber and optimization of the capture process, three automated captures of 25 cycles each were run. Each of those three captures used a fresh membrane adsorber.
Table 1: Optimized volumes and flow rates for each cycle step, with the same method used for experiments for the Sartobind Rapid A Nano and Mini prototype devices; MV = membrane volume. 1 without loading 2 depending on binding capacity and product concentration.
Capture Scale-Up to Technical Scale: Capture scale-up was performed using a Sartobind Rapid A mini prototype (4-mm bed height, 10 mL total membrane volume) provided by Sartorius Stedim Biotech. An ÄKTA pilot 400 chromatography system was used, controlled by Unicorn 5.3.1 software, both from Cytiva. Again, the membrane adsorber and the chromatography system were prepared and stored as recommended by their suppliers. All preparations, process steps, and buffer conditions from the laboratory-scale process were applied.
After three preliminary tests, one automated, scaled-up capture of 16 cycles was performed. To ensure success despite the short optimization phase, a safety margin was applied in which the volumes for elution and CIP were increased by a factor of ~20. All preliminary and scaled-up tests used the same membrane adsorber.
Analytics: Every important process parameter for performance evaluation in this case — UV-absorbance signal, pressure, flow rate, conductivity, and pH — was evaluated using the Unicorn software, with calculations by Microsoft Excel and graphical representations made using Origin 2022 software from OriginLab. Elution product yields were determined by UV absorbance measurement at 280 nm with an Infinite 200 Pro plate reader from Tecan Life Sciences. Because that method gave a total protein determination, the results were corrected by factoring for impurities that remained in each elution.
To determine the amount of impurities from which those could be derived, purity was determined by separating proteins using hydrophobic-interaction chromatography (HIC) using a Cytiva column and a program designed by employees of Bibitec GmbH & Co. KG. Chinese hamster ovary (CHO) host-cell protein (HCP) removal was determined with a third-generation CHO HCP ELISA kit from Cygnus Technologies. CHO-cell DNA was determined with digital polymerase chain reaction (dPCR) using a QIAcuity CHO resDNA Quant kit after extraction using a QIAamp DNA Mini kit (both from Qiagen).
Results and Discussion
Experimental Foundation and Idea: The existing capture method based on column material for the target Fc-fusion protein performs very well on an industrial scale, purifying up to 200 L of culture supernatant. Thus our experimental aim was to project a capture method using a Sartobind Rapid A membrane adsorber to that same scale for performance comparability analysis. Table 2 overviews our projection for comparison between the column- and membrane-based capture methods, including assumptions derived from the existing process.
Table 2: Overview of parameters and assumptions needed to compare and project a developed membrane-based capture method against an existing column-based process at 200-L scale; DBC = dynamic binding capacity. 1 Calculated volume to meet column-based process duration 2 own evaluation.
The existing column-based capture requires two cycles and is associated with two days of work. Assuming linear scalability and based on the Sartobind Rapid A calculation presented in Table 1, a 200-mL Sartobind Rapid A membrane adsorber is required. It requires 16 cycles to complete the entire process in the same amount of time. Unlike what is now common for mAbs (4), the target Fc-fusion-protein is expressed at <0.5 g/L. Because of that comparably low titer, a high volume per cycle must be loaded onto the membrane adsorber until it reaches capacity. In conjunction with the necessary flow rate, that volume requirement leads to loading times of >25 minutes, which somewhat limits the time gain from using a membrane adsorber. Note that, according to the manufacturer, standard product concentrations can allow for cycle times of <10 minutes.
The low concentration of the Fc-fusion protein also entails a low product yield, which is why the required number of cycles tested here is significantly lower than the maximum number of >200 cycles tested by the manufacturer (11). That lower number of cycles compensates for the longer cycle duration of the membrane-based capture method. The developed method could be accelerated by increasing Fc-fusion protein concentration in the culture supernatant or by choosing a larger membrane adsorber from the Sartobind Rapid A portfolio. Note that increases to membrane size are limited by associated increases in flow rates. Even with a 200‑mL Sartobind Rapid A membrane adsorber, flow rates of 2 L/minute are required for the capture method developed herein.
Nevertheless, from our projection, all experiments should cover a minimum of 16 cycles to capture the Fc-fusion protein with a Sartobind Rapid A membrane adsorber with comparable performance to the existing capture method based on column material.
Investigation of Capture Performance at Small (Laboratory) Scale: Calculations revealed that capture performance must be studied over at least 16 cycles to ensure projection of the entire process to 200-L scale. Laboratory-scale experiments used the Sartobind Rapid A Nano membrane adsorber, which holds a volume of 1 mL. With about 2 L of culture supernatant, 25 cycles were performed according to conditions and assumptions from Tables 1 and 2. To evaluate the functionality of this small-scale capture, we examined the performance of various process parameters throughout all cycles (Figures 1a and 1b).
The chromatograms are congruent for all 25 cycles (Figure 1a). After equilibration (not included in the chromatograms), a large flowthrough occurs in each cycle, indicating strong impurity removal. That observation is underlined by a final sample purity of nearly 92% on average, confirming successful removal of impurities. In fact, a 7% higher purity is obtained by capture with Sartobind Rapid A compared with the resin-based benchmark process.
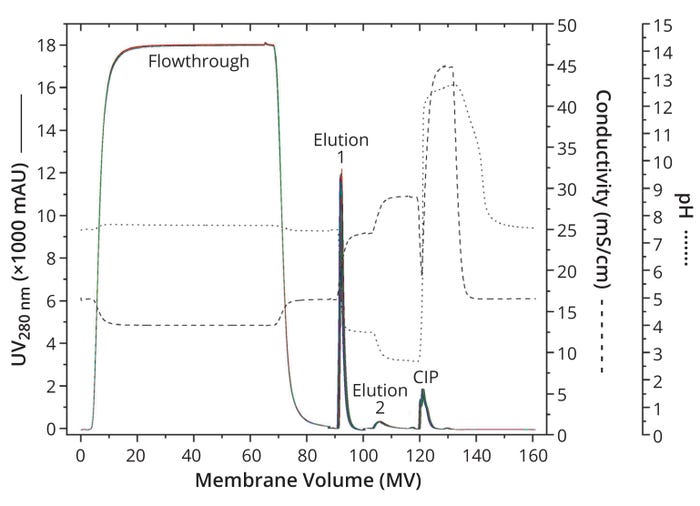
Figure 1a: Overlay of chromatograms for 25 capture cycles at laboratory scale; UV absorbance at 280 nm for all cycles are distinguished by color and shown as a solid line. For better orientation, the conductivity curve is indicated as a dashed line and the pH curve as a dotted line; CIP = clean in place.
Washing followed sample application until the UV 280-nm signal dropped below 10 mAU. At this step, the highest flow rate of the whole process was used, and the highest pressures were obtained (Figure 1b). Note that no significant pressure increase was observed over all 25 cycles, indicating a lack of membrane fouling and proving the robustness of the process. Moreover, the maximum recorded pressure value of 0.23 MPa is well shy of the 0.4 MPa pressure limit, demonstrating that many more than 25 cycles can be performed based on the method described herein (Table 1).
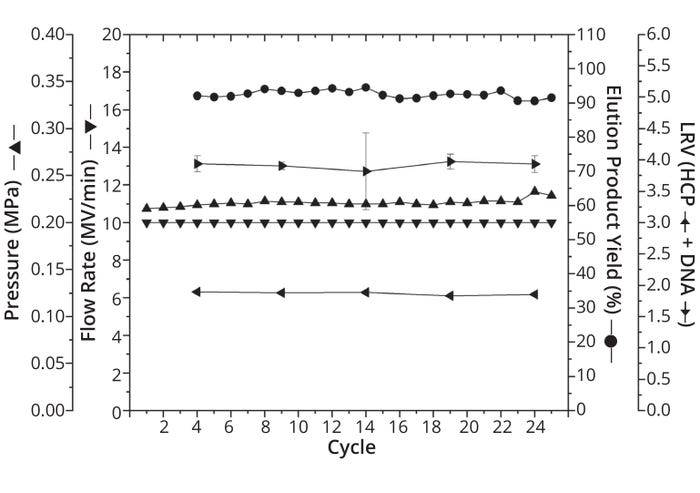
Figure 1b: Development of different process parameters over the course of 25 capture cycles with the following parameters plotted for each cycle: % yield in elution (indicated as points), highest flow rate in MV/min (indicated as downward-pointing triangles), maximum pressure in MPa (upward-pointing triangles), log reduction value (LRV) of Chinese hamster ovary (CHO) host-cell proteins (HCP) (leftward-pointing triangles), and LRV of CHO DNA (rightward-pointing triangles). Because of limited assay volume, LRV data were examined for only five cycles. Except for pressure and flow rate, all parameters were measured as triplicate determinations (n = 3). Standard deviations are marked as error bars.
In alignment with the resin-based benchmark process, the target molecule was eluted by a two-step pH-gradient elution. Uniformity across all cycles — and therefore functionality of capture with the membrane adsorber — is particularly evident when you consider the average product yield in elution: ~92.5% ± 1.2%. That result is in line with the yield obtained by the manufacturer of classical mAbs, which ranges from 80% to 100%. And it indicates that Sartobind Rapid A membrane adsorbers can be a powerful tool for capture of both mAbs and Fc-fusion proteins.
That conclusion is substantiated further by an average log reduction value (LRV) for CHO HCP of 1.90 ± 0.02 and for CHO DNA of ~3.90 ± 0.05 across all cycles. LRV of CHO HCP was comparable to that of the resin-based benchmark process, whereas CHO DNA removal was 15% higher with the resin-based process. Nevertheless, our data reveals similar results for both materials, meaning that they perform comparably. We find that to be impressive considering the low product concentration and thus enormous volume of supernatant that had to be loaded onto the membrane adsorber — increasing the burden of impurities in that process.
The CIP peak again demonstrates comparability across all 25 cycles. We conclude that CIP using 0.2 M NaOH prevents fouling of the membrane adsorber. However, that is true only as long as CIP follows each cycle.
Our data show successful capture of Fc-fusion protein using a Sartobind Rapid A Nano membrane adsorber at laboratory scale over 25 cycles. Throughout all cycles, neither yield nor purity deviated; both were comparable to the benchmark resin-based process. Future experiments could elucidate whether the ≥200 cycles tested by the membrane manufacturer can be achieved (11). But the consistent results and comparable performance are striking considering the fact that low product concentration in the culture supernatant necessitated loading of a comparatively large volume onto the membrane adsorber in each cycle (bringing high levels of impurities). Thus, the results of this small-scale study indicate that scale-up to a 200 L is highly feasible.

Small-scale membrane chromatography system used in this study.
Scalability and Capture Performance at Technical Scale: A scale-up factor of 10 was applied to a Sartobind Rapid A Mini membrane adsorber with a 10-mL membrane volume. Based on previous calculation (Table 2), we needed to investigate at least 16 cycles to assess whether a transfer to 200-L production scale would be feasible. Therefore, these experiments used about 16 L of culture supernatant. To evaluate the linear scale-up, various process parameters were examined across all 16 cycles (Figures 2a and 2b).
As at small scale, the chromatograms are consistent for all 16 cycles, indicating a robust process over 16 cycles. Differences in flowthrough can be attributed to differences in the feed rather than membrane performance. The high volume made it possible to prepare the culture supernatant in two steps, with the second half added to the sample bag after the seventh cycle, resulting in an increased Fc-fusion protein concentration of ~30%. Assuming that other components of the culture supernatant increased at the same rate, the higher signal during flowthrough can be explained, especially because the signal increased by ~30% as well.
Consistent cycle-to-cycle performance is indicated by a consistent and impressively high yield of 98.3% ± 2.0%. Additionally, LRVs remained consistent across all cycles at 2.00 ± 0.08 for CHO HCP and 4.12 ± 0.07 for CHO DNA. Both LRVs are comparable to the benchmark resin-based process and in the same order of magnitude as the small-scale capture results.
However, the pressure acting on the membrane increases from 0.07 MPa to 0.12 MPa across 16 cycles, which represents an increase of >70% relative to the initial value. Note that the first significant increase of 0.02 MPa occurs after the seventh cycle, when the second half of the culture supernatant was added. Such increases might stem from increases in supernatant concentration (Figure 2a). The resulting increase in stress on the membrane adsorber might have reached a point at which the CIP was too weak to remove all impurities — a possibility that is supported by the relative height of the CIP peaks. However, the pressure remained well below the 0.4-MPa limit, so its increase did not affect the functionality, performance, or consistency of the scaled-up capture. Therefore, we deem that a transfer of the Fc-fusion protein capture process to a Sartobind Rapid A membrane adsorber at the production scale of 200 L remains feasible.
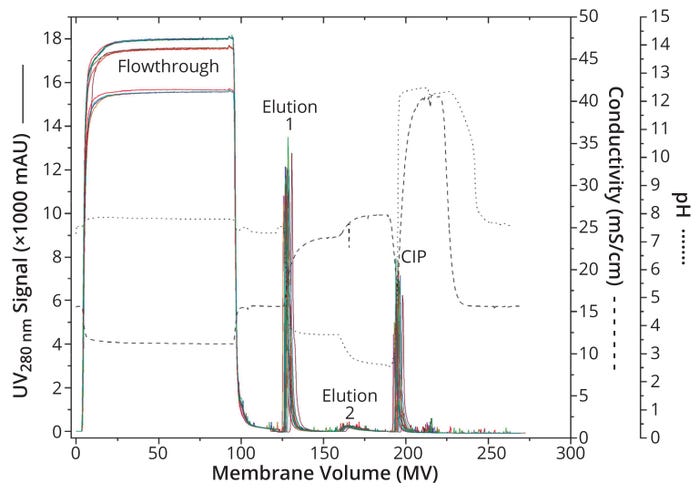
Figure 2a: Chromatogram overlay of 16 cycles at technical scale; absorbance at 280 nm for all cycles is distinguished by color and shown as a solid line. For better orientation, the conductivity curve is indicated as a dashed line and the pH curve as a dotted line.
Another important aspect to mention is that desired linearity cannot be fully maintained through scale-up. When considering the volumes required for variable steps of each cycle (data not shown), we found scaling factors of 9.5–13.0 for the wash, elution 2, and CIP steps when comparing laboratory- and technical-scale operations. In those steps, scale-up seems to be linear. For elution 1, however, our study shows a scaling factor of nearly 22, which is related to the increased tailing of the peak during the scaled-up capture method. With longer development times, that phenomenon could have been circumvented. Nevertheless, the assumption of linearity in scale-up is a good orientation for process development, when fine details must be considered individually for each process.
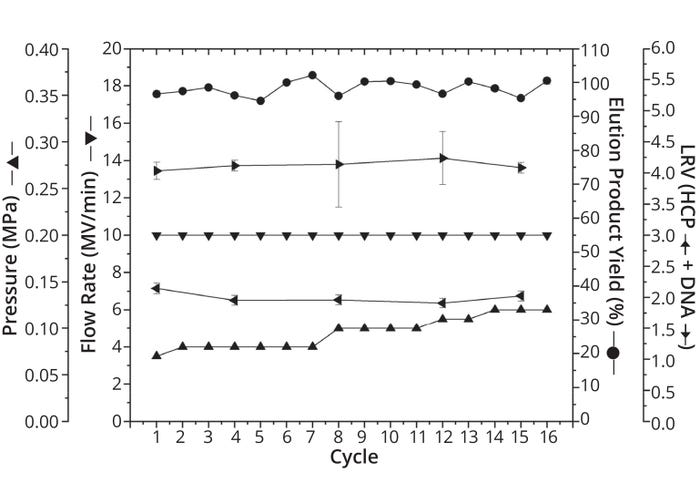
Figure 2b: Critical process parameters over 16 cycles from technical scale, with the following parameters plotted for each cycle: % yield in elution (indicated as points), highest flow rate in MV/min (downward-pointing triangles), maximum pressure in MPa (upward-pointing triangles), log reduction value (LRV) of Chinese hamster ovary (CHO) host-cell proteins (HCP) (leftward-pointing triangles), and LRV of CHO DNA (rightwardpointing triangles). Because of limited assay volume, LRV data were examined in only five cycles. Except for pressure and flow rate, all parameters were measured as triplicate determinations (n = 3). Standard deviations are marked as error bars.
In terms of uniformity and consistency, technical-scale performance aligned with that at laboratory scale, manufacturer data, and the benchmark resin-based capture method — demonstrating a successful scale-up. Conditions selected for capture of this Fc-fusion protein using the Sartobind Rapid A membrane adsorber appear to be projectable and transferable to 200-L production scale.
Conclusion and Outlook
Comparison and Choice of the Best Option: It has been demonstrated that the capture of an Fc-fusion protein at 200-L production scale seems possible within two days using both conventional chromatography resin and a Sartobind Rapid A membrane adsorber. Moreover, the processes performed comparably within the range of typical values for protein A capture. To determine which method is the better choice, we compared 12 critical process parameters (Table 3). For 10 of those parameters, the membrane adsorber shows identical or superior results.
Table 3: Comparing critical process parameters for Sartobind Rapid A membrane adsorber and a conventional protein A resin; DBC = dynamic binding capacity, HCP = host-cell protein, LRV = log reduction value. 1 percentages compared with each other 2 already projected to 200-L scale.
Particularly interesting is the dynamic binding capacity (DBC) at 10% flowthrough, which for both materials is at least 30 g/L. That result indicates technological improvements made in recent generations of membrane adsorbers (6). Another parameter to highlight is the 15-fold higher loading speed and/or 15-fold shorter residence time, confirming a key advantage of membrane-based chromatography over resin-based chromatography that already has been shown for mAbs (5, 13). The Sartobind Rapid A membrane adsorber thus performs comparably with molecules of different affinity for the protein A ligands. Finally, the membrane adsorber coped well with application of high sample volumes and resulting increases in contamination load without loss of performance.
Cost reduction mainly results from significantly lower chromatography media requirements in the case of membrane adsorbers. Those requirements — and therefore smaller devices — offer an additional process-handling advantage by omitting column packing and cleaning. But simpler process handling cannot be generalized because buffer requirements concomitantly increase by ~30%.
Admittedly, time and cost calculations were greatly simplified in this work. Not counting the cost of regeneration itself, we calculated that the remaining costs equalize after the resin material has been regenerated about five times. A five-fold regeneration is also quite realistic without performance loss (14).
Taking into account the advantages of increased productivity, applying single-use capsules, lowered risk of in-process contamination, and reduced validation effort, our data indicate that capturing the Fc-fusion protein with a Sartobind Rapid A membrane adsorber should lead to process intensification. Further calculations have revealed that the real advantage of this membrane adsorber comes into play with significantly higher product quantities. Increasing Fc-fusion protein concentration by a factor of 10 could reduce the time required for the entire process by three days at roughly similar costs.
Our work shows that the capture method developed and investigated herein can be operated with both types of chromatography material and parameters acceptable for production. We show that membrane-based chromatography has taken a decisive step forward. However, which material ultimately represents the better option must be evaluated individually for each downstream process.
Acknowledgments
This work originates from author Knoll’s master thesis at the Institute of Biotechnological Process Engineering in the Department of Engineering and Mathematics at Bielefeld University in Germany. Membrane adsorbers were provided by Sartorius Stedim Biotech. Knoll was a student and thus at no time was an employee of or sponsored by Sartorius AG. Thanks go out to all who made the cooperation possible — in particular, Dr. Ricarda Busse and Marius Lechner at Sartorius and Dr. Dirk Lütkemeyer and colleagues at the university.
References
1 Weiner LM. Fully Human Therapeutic Monoclonal Antibodies. J. Immunother. 29(1) 2006: 1–9; https://doi.org/10.1097/01.cji.0000192105.24583.83.
2 Liu L. Antibody Glycosylation and Its Impact on the Pharmacokinetics and Pharmacodynamics of Monoclonal Antibodies and Fc-Fusion Proteins. J. Pharmaceut. Sci. 104(6) 2015: 1866–1884; https://doi.org/10.1002/jps.24444.
3 Köhler G, Milstein C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature 256, 1975: 495–497; https://doi.org/10.1038/256495a0.
4 Huang Y-M, et al. Maximizing Productivity of CHO Cell-Based Fed-Batch Culture Using Chemically Defined Media Conditions and Typical Manufacturing Equipment. Biotechnol. Progr. 26(5) 2010: 1400–1410; https://doi.org/10.1002/btpr.436.
5 Brämer C, et al. Membrane Adsorber for the Fast Purification of a Monoclonal Antibody Using Protein A Chromatography. Membranes 9(12) 2019: 159; https://doi.org/10.3390/membranes9120159.
6 Boi C, Dimartino S, Sarti GC. Performance of a New Protein A Affinity Membrane for the Primary Recovery of Antibodies. Biotechnol. Progr. 24(3) 2008: 640–647; https://doi.org/10.1021/bp0704743.
7 Boi C. Membrane Adsorbers as Purification Tools for Monoclonal Antibody Purification. J. Chromatogr. B 848(1) 2007: 19–27; https://doi.org/10.1016/j.jchromb.2006.08.044.
8 Bolton GR, Mehta KK. The Role of More Than 40 Years of Improvement in Protein A Chromatography in the Growth of the Therapeutic Antibody Industry. Biotechnol. Progr. 32(5) 2016: 1193–1202; https://doi.org/10.1002/btpr.2324.
9 Ghosh R. Protein Separation Using Membrane Chromatography: Opportunities and Challenges. J. Chromatogr. A 952(1–2) 2002: 13–27; https://doi.org/10.1016/s0021-9673(02)00057-2.
10 Nadar S, et al. Intensified Downstream Processing of Monoclonal Antibodies Using Membrane Technology. Biotechnol. J. 16(3) 2021: 2000309; https://doi.org/10.1002/biot.202000309.
11 Grünberg M, et al. Scalable, Robust, and Highly Productive Novel Convecdiff Membrane Platform for mAb Capture. Membranes 12(7) 2022: 677; https://doi.org/10.3390/membranes12070677.
12 Vogel JH, et al. A New Large-Scale Manufacturing Platform for Complex Biopharmaceuticals. Biotechnol. Bioeng. 109(12) 2012: 3049–3058. https://doi.org/10.1002/bit.24578.
13 Ghose S, Hubbard B, Cramer SM. Binding Capacity Differences for Antibodies and Fc-Fusion Proteins on Protein A Chromatographic Materials. Biotechnol. Bioeng. 96(4) 2007: 768–779; https://doi.org/10.1002/bit.21044.
14 Pathak M, Rathore AS. Mechanistic Understanding of Fouling of Protein A Chromatography Resin. J. Chromatogr. A 1459, 2016: 78–88; https://doi.org/10.1016/j.chroma.2016.06.084.
Corresponding author Florian Knoll, MSc, is a downstream process specialist at Bibitech GmbH & Co. KG in Uetersen Germany; [email protected]. Marius Lechner, MSc, is a chromatography applications specialist, and Ricarda Busse, PhD, is global product manager for membrane chromatography at Sartorius Stedim Biotech. Dirk Lütkemeyer, PhD, is a professor in the Institute of Biotechnological Process Engineering at Bielefeld University in Germany.
You May Also Like






