- Sponsored Content
- Biosimilars
- Product Characterization
Building Orthogonality into Biosimilar TestingBuilding Orthogonality into Biosimilar Testing
April 13, 2020
Sponsored by BioPharmaSpec
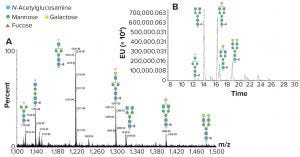
Figure 1: N-glycan analysis using peptide mapping and fluorescent tagging; (a) shows peptide-mapping signals observed for an IgG glycopeptide, and the region containing doubly charged signals is shown (only the glycans are shown for simplicity). A relatively high abundance of truncated glycan species can be seen (marked by an asterisk). (b) is a chromatographic profile of fluorescently tagged released N-glycans. The truncated species seen in Figure 1a are either absent or significantly reduced, indicating they come from in-source fragmentation processes in the peptide mapping experiment.
Both the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) have developed regulatory guidelines on biosimilars (1, 2). These comprehensive documents provide clear guidance on what the agencies expect from structural characterization studies. Using state-of- the-art instrumentation and techniques is expected, but the use of orthogonal techniques in structural comparability assessments is also required. The application of orthogonal analytical techniques will provide a firm structural foundation to claims of biosimilarity. Characterization methods verifying and supporting conclusions drawn from other techniques will provide a robust package of structural data in which both manufacturers and regulatory authorities can be confident. Below, the author describes three examples of orthogonality within required analyses, each examining different aspects of protein or glycoprotein structure.
Glycan Analysis
Data on glycan structure can be generated in different ways, but it is important to understand the limitations of the methods used. For example, as part of an investigation into a glycoprotein’s structure, peptide mapping should be performed. Peptide mapping is the identification of peptides (through the use of liquid chromatography–mass spectrometry, LC-MS) from a series of specific proteolytic digestions of a protein. Masses of detected peptides should match theoretically predicted masses, providing confidence in several aspects of the protein’s structure (e.g., sequence, intactness, and presence or absence of posttranslational modifications). Basic peptide mapping is insufficient for confirming a primary amino-acid sequence unless specific evidence for each amino acid is determined and other techniques are used to eliminate ambiguities.
Peptide mapping can detect glycopeptides. However, energy applied during ionization (within a mass spectrometer) can lead to some fragmentation of glycan structures, producing a profile that contains artificially generated truncated species as well as glycans that are naturally present on the glycopeptide (Figure 1a). Because glycopeptides have a lower ionization efficiency than peptides, it is unlikely that all glycopeptides will be detected (with minor species not being observed above a background).
There is no doubt peptide mapping is very important for assessing glycopeptides and provides important structural evidence for their presence and nature (and the level of glycosylation site occupancy), but a full assessment of the glycans requires release and analysis in specifically designed experiments (such as on-line LC–MS analysis of fluorescently labeled released glycans). This technique does not present the same issues as peptide mapping because glycan species are separated, and retention time and peak area of the fluorescently labeled species are used to identify and provide relative quantitation (Figure 1b).
Mass spectrometric assessment of the eluate then provides intact mass information and structural information for each released oligosaccharide through fragmentation, the combination allowing identification of each component. The fluorescently labeled data will serve to verify and extend the conclusions drawn from peptide mapping data. This is particularly useful because glycopeptides tend to produce minimal peptide fragmentation in a basic mapping study. Thus, the components observed in the fluorescent data can be used to support the glycopeptide mass assignments made from the mapping data.
Assessments of C-Terminal Lysine on MAbs
The C-terminal lysine on the heavy chain of monoclonal antibodies (MAbs) is a common posttranslational modification (3). It can be detected readily in a peptide map with the presence of C-terminal heavy-chain peptides with and without lysine. Fragment-ion information generated concurrently in a peptide mapping exercise (e.g., by quadrupole time-of-flight (Q-TOF) MS) supports this assignment.
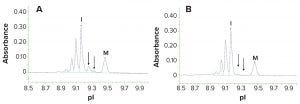
Figure 2: Imaged capillary isoelectric focusing (icIEF) electropherogram of an IgG before (a) and following (b) treatment with carboxypeptidase B to remove C-terminal lysine from the heavy chain; lysine containing components (arrowed) are absent following treatment with the enzyme. The components at more acidic pI are deamidated species. The peaks labelled “I” and “M” are intact monoclonal antibody with no heavy chain C-terminal lysine and a pI marker, respectively.
Imaging capillary isoelectric focusing (icIEF) can be performed as an orthogonal identification of C-terminal lysine on MAbs. In this technique, charged isoforms of the sample migrate to their isoelectric points (pI). Because lysine is a basic residue, species that have the C-terminal lysine migrate to a more basic pI than those that do not. MAbs contain two heavy chains, so icIEF evaluation can produce three peaks: MAb with no C-terminal lysine, MAb with a C-terminal lysine on one heavy chain, and MAb with a C-terminal lysine on both heavy chains. Pretreatment of a sample with enzyme carboxypeptidase B removes remaining lysine residues at the heavy chain C-terminus and eliminates the “with lysine” signals when run in a second icIEF experiment. This not only confirms the C-terminal lysine modification, but also serves to identify the nature of basic peaks seen in the original icIEF analysis (Figure 2).
Higher-Order Structure Analysis
The final example of the use of orthogonality is in the area of higher-order structure (HOS) analysis. Assessments of secondary and tertiary structure are regulatory requirements and are examined using techniques that assess different aspects of such HOS. Such methods include circular dichroism (CD), Fourier-transform infrared (FTIR) spectroscopy, and nuclear magnetic resonance (NMR). Each technique probes different aspects of HOS and generates data according to the nature of the technique used. Each method has its own area of strength (e.g., alpha helix or beta sheet detection).
HOS analysis across these various techniques is a case of the sum being more than the parts. A combination of the data obtained provides a very robust orthogonal assessment of secondary and tertiary structure and an in-depth assessment of comparability. In a structural study — regardless of whether it is an assessment of biosimilarity — data generation in this way is becoming the regulatory expectation in terms of investigating HOS.
In summary, use of orthogonal techniques for structural characterization not only serves to provide a robust overall package of information about the nature of a product, but it “cross-verifies” conclusions drawn from within different data sets, thereby adding to the robustness of the package as a whole.
References
1 Guidance for Industry: Development of Therapeutic Protein Biosimilars: Comparative Analytical Assessment and Other Quality-Related Considerations. US Food and Drug Administration: Rockville, MD, May 2019.
2 Guideline on Similar Biological Medicinal Products Containing Biotechnology-Derived Proteins as Active Substance: Quality Issues (Revision 1). EMA/CHMP/BWP/247713/2012. European Medicines Agency: Amsterdam, Netherlands, May 2014.
3 Beyer B, et al. Microheterogeneity of Recombinant Antibodies: Analytics and Functional Impact. Biotechnol. J. 13, 2018: doi.10.1002/biot.201700476.
Richard L. Easton is technical director of structural analysis at BioPharmaSpec;
[email protected].
You May Also Like






