Detection and Clearance of Viruses in the Biopharmaceutical IndustryDetection and Clearance of Viruses in the Biopharmaceutical Industry
December 17, 2019
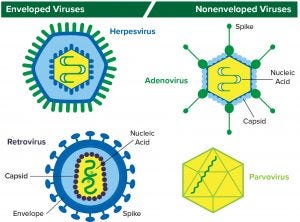
Figure 1: Structure of enveloped viruses (represented by herpesvirus and retrovirus) and nonenveloped viruses (represented by adenovirus and parvovirus) www.istockphoto.com
Viral contamination is a common threat to all animal- and human-derived biopharmaceuticals. This type of contamination can affect any part of a bioproduction process, so biomanufacturers need to perform viral testing studies and incorporate viral clearance methods into their processes.
Viral contaminants can come from cell lines (e.g., endogenous retroviruses) or from adventitious (e.g., mycoplasma) introduction during drug manufacturing. Virus testing of master cell banks (MCBs), working cell banks (WCBs), end-of-production cell banks, and bulk unprocessed harvest material is called for in guidance documents including Q5A from the International Council on Harmonisation of Technical Requirements for the Registration of Pharmaceuticals for Human Use (ICH) (1). Regulators also recommend that manufacturers source raw materials appropriately and use effective methods for demonstrating viral clearance — and to use a risk-based approach when doing so.
Thus, viral detection and clearance studies must be performed to demonstrate clearance of viruses known to be related to a process. By illustrating the ability of the process to clear specific and unspecified viruses, such studies also test the robustness of a process for removal of potential adventitious viruses that could enter a production process through a number of routes.
Test methods applied for virus detection will be examined herein along with viral clearance processes used. In addition, those methods should be researched to determine their potential and limitations for viral clearance. Because it is not practical to attempt to cover all categories of biopharmaceutical products and cell lines, I use Chinese hamster ovary (CHO) cells as an example when referring to common viruses found.
Background
Some viruses can cause morphological changes to infected cells and are somewhat easy to detect; others do not affect the morphology of infected host cells and thus are much more difficult to detect (2). Moreover, a product can be contaminated by adventitious and/or endogenous viruses. According to the World Health Organization (WHO), adventitious viruses are those “that have been unintentionally introduced into the manufacturing process of a biological product” (3). They can come from raw materials, cell culture media, and cell lines as well as other sources such as equipment, personnel, and the environment. By contrast, endogenous viruses are those “whose genome is present in an integrated form in a cell substrate. Endogenous viruses are present in the genome of the original animal from which the cells were derived. They may or may not encode an intact or infectious virus” (3).
A virus is essentially an infectious agent that is capable of infecting a host cell and to copy itself. Viruses consist of nucleic acid (DNA or RNA) in a protein coat/capsid. Those with an outer lipid layer are referred to as enveloped viruses; those without one are classified as nonenveloped or naked viruses. DNA-based herpesviruses and RNA-based retroviruses are examples of enveloped viruses; DNA-based adenoviruses and RNA-based parvoviruses are examples of nonenveloped viruses (Figure 1).
Product Safety: No single strategy can ensure the safety of all biological products. Analytical limitations make it impossible to demonstrate absolute viral absence. Ensuring that a finished product is free from viral contamination does not depend exclusively on testing for the presence of viruses. It also requires viral clearance methods to inactivate and/or remove them (4).
As mentioned above, virus contamination can arise from a cell line itself or from adventitious introduction during bioprocessing. Several complementary methods can be used to tackle both types: testing product at certain points during production and processing for the presence of viral contaminants, testing cell lines and raw materials for endogenous viruses, and challenging a process and testing its ability to remove them.
In some cases, detection systems are not adequately sensitive to identify viral contaminants, and both adventitious and endogenous viruses can escape detection. That can happen for several reasons such as restricted assay sensitivity (5). Also, the persistent threat of new and emerging viral contaminants is inevitable.
Biopharmaceutical product manufacturers and suppliers have taken a number of steps to avert viral contamination. Examples include use of well-established and -characterized cell lines and chemically derived rather than animal-sourced materials, establishment of risk mitigation and control procedures for sourcing raw materials and excipients, and application of bioprocess methods that prevent viral contamination (6).
Virus Detection
A number of methods can be used to detect endogenous and adventitious viruses. Biopharmaceutical manufacturers must select and design the most appropriate virus detection test methods for use across a manufacturing process, from initial cell lines up to the final-product stage, including raw materials and bulk drug-substance harvest. In fact, regulators require that testing be carried out at every stage of a biomanufacturing process (7).
When you are developing and deciding on a virus detection/testing strategy, it is important to consider the risk of contamination from endogenous and adventitious viruses both in starting materials and in the final product. Because no particular assay or approach can detect all viruses, a number of orthogonal methods (both general and specific) are required for virus detection (7). Test methods can be divided into three broad categories: species-specific assays for known potential contaminants, general nonspecific methods, and retrovirus assays.
Those methods detect viruses in different ways. Some assays measure viral contamination in test cells, some search for viral proteins or particles, and others look for viral markers in addition to the presence of viral genomes (8). Here are examples of each:
viral contamination in test cells — in vitro and in vivo assays
viral proteins — enzyme assays for reverse transcriptase (RT) in retroviruses
viral particles — electron microscopy (EM)
viral markers — polymerase chain reaction (PCR) assays.
Specific and Nonspecific Methods: General nonspecific assays considered to be the industry standard for detecting adventitious viruses include transmission electron microscopy (TEM) and infectivity testing using animals (in vivo) and cell cultures (in vitro). The most commonly used species-specific (target-specific) assay is quantitative PCR (qPCR). Even though nonspecific assays can detect a broad range of viruses, some types can go undetected because of diverse viral physiology and limitations in assay sensitivity. By contrast, species-specific methods generally are highly sensitive but can detect only known/predefined target(s). Furthermore, PCR-based assays can distinguish between infectious and noninfectious viruses. Thus, target-specific methodologies often are used as tools to support viral contamination investigations after positive results from general screening assays (9).
Retrovirus Assays: In addition to the above techniques, regulatory agencies request that companies use EM when testing for the presence of retrovirus in cells and that product sponsors quantify retrovirus-like particles in bulk-harvests, such as those from rodent cells (e.g., CHO cells) (7). All the standard methods have limitations such as long assay times, detection only of specific target viruses, and an inability to identify unknown viruses. In addition, they cannot detect viruses that do not elicit cytopathogenic effects (CPEs), which are structural changes in host cells they have invaded. Consequently, next-generation sequencing (NGS) technology — also known as massive parallel sequencing (MPS) or deep sequencing — has been adopted in recent years to address those limitations in detecting adventitious viruses. NGS is principally a high-throughput DNA sequencing method capable of sequencing millions of nucleic acids in a sample. This powerful tool has the flexibility to sequence a range of nucleic acids and can detect a wide range of both new and unknown viruses. NGS also offers the impressive ability to sequence more than 40 human genomes in a single day — compared with the 13 years it took to sequence the first human genome years ago (10, 11).
Even though NGS/MPS is a relatively new technology, regulatory agencies are beginning to recognize its benefits. For example, the European Pharmacopoeial Commission’s ninth edition issued a guidance document that refers to the use of deep sequencing as an alternative to in vivo or as a supplement/alternative to in vitro tests for adventitious virus detection (12).
Viral Clearance
No single test can detect all viruses, and all methods require a minimum level of viruses to be detected. Some methods measure markers of infection, providing results long after infection occurs. Hence, ensuring that a product is free from viral contamination depends on ensuring that a process can inactivate or remove viruses (through viral clearance validation studies) as well as testing for their presence. Such an approach is the only way to ensure that biopharmaceuticals will be free from viral contamination and safe for human use (13).
When designing viral-clearance validation studies, it is important to recognize which viruses can possibly contaminate a product and enter its manufacturing process. Nearly all cell genomes include some retrovirus sequences. Indeed, some cell lines commonly used for biopharmaceutical production (e.g., CHO and murine cells) have been found to release retrovirus-like particles. Although the retroviruses found in CHO cells have been determined not to be infectious (14), those found in murine cells could be (15). Other viruses that replicate in CHO cells include vesivirus, reovirus, mouse minute virus (MMV), and Cache Valley virus (CVV), to name a few (16, 17).
It is essential to test the ability of clearance methods to remove viruses in bioprocessing, so targeting a wide range of physicochemical properties is advised. The US Food and Drug Administration (FDA) categorizes viruses used for clearance studies into three types: specific model viruses, nonspecific model viruses, and product-/process-relevant viruses (1). Using nonspecific model viruses with different physicochemical properties helps a company determine the capability of its manufacturing process to inactivate and/or remove viruses in general.
Relevant viruses are those that are known to potentially contaminate cell substrates or other materials used in a biomanufacturing process. These viruses should be removed by a viral inactivation/removal process. A specific model virus is one that is closely related to a relevant virus (same family or genus) and has similar chemical and physical properties. Specific viruses are used when a relevant virus is unavailable, e.g., because it cannot be laboratory-grown at required levels for testing (1).
Common examples of viruses used to represent a wide range of physicochemical properties include small, nonenveloped viruses such as a poliovirus or an animal parvovirus; large DNA viruses such as herpesvirus; and large enveloped RNA viruses such as murine retrovirus or parainfluenza virus. In viral clearance studies, companies deliberately spike samples of such viruses at different stages throughout a down-scaled manufacturing process — and analyze the ability of that process to inactivate or remove them by the method of choice. Doing so will help identify which methods are effective and measure their effectiveness. It is important to select methods and processes that closely resemble the production-scale process they intend to represent.
Not every single step of a biomanufacturing process needs to be tested. Only steps that are known to clear (inactivate/remove) viruses should be included in a validation study (13). Sample materials should be spiked with high enough levels of viruses to challenge the process but not enough to alter the sample material (18). Typical volumes used are >10% of a virus load in the sample to be tested. In addition, a process typically will be evaluated and tested under both normal and abnormal operating conditions (e.g., high temperatures, pH levels, and agitation) to validate robustness.
To prevent introduction of viruses into a production facility, viral clearance studies generally are performed in specialized virological laboratories in the form of scaled-down versions that mimic a biomanufacturing process. Ideally, whether reduction of viral infectivity is achieved by inactivation or removal should be noted. Results obtained from scaled-down methods are adjusted and compared with parameters (e.g., pH, protein concentration, and temperature) of a large-scale process. Test results should be quantitative and have suitable sensitivity and reproducibility. Common methods used for these studies include quantal endpoint titrations such as tissue culture infectious dose (TCID50) assays and qualitative nucleic-acid amplification methods such as PCR.
All methods are susceptible to error. For example, even though PCR is recognized as an excellent method with high sensitivity for detecting viral genomes, unfortunately it also detects “inactivated” viruses, which can lead to false-positive results and thus misrepresent a potentially valid viral-inactivation step. Therefore, PCR techniques tend to be used to test viral “removal” methods rather than those for inactivation.
It is important to consider several factors when interpreting results before concluding whether a particular clearance method is acceptable (1): e.g., test viruses used, log reduction values (LRVs) achieved, kinetics of viral inactivation, methods of inactivation/removal used, and assay limit of detection (LoD).
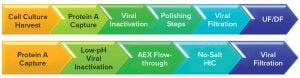
Figure 2: Amgen’s (top) and Biogen’s (bottom) monoclonal antibody downstream processing approaches include their chosen viral clearance methods (18); UF/DF = ultrafiltration/diafiltration, AEX = anion-exchange chromatography, HIC = hydrophobic-interaction chromatography
Viral clearance methods generally are performed as part of downstream processing (Figure 2). However, because viral contamination also can occur in upstream production areas — particularly in bioreactors — many biomanufacturers choose to apply viral clearance screening and mitigation techniques to the upstream side, particularly for cell culture media and components (17).
A number of viral clearance methods currently are used in biopharmaceutical manufacturing for viral inactivation or removal. For example, high-temperature–short-time (HTST) treatment and UV-C (ultraviolet-C light at 200- to 280-nm wavelengths) are used in upstream processing areas. In the former method, a protein solution is subjected to continuous flowrates and exposed to high temperatures for short periods, typically 70–75 °C for 30 seconds. The UV-C method is effective for inactivating nonenveloped viruses (19). Examples of viral inactivation methods commonly used in downstream areas include solvent/detergent treatment and low-pH (acidic) viral inactivation, which effectively targets enveloped viruses. Other less common methods used for inactivating enveloped viruses include microwave heating, irradiation (UV and gamma), pasteurization, and HTST.
Typically, each viral clearance method is tested separately, after which the amount of clearance obtained from an entire process is calculated from a sum of the results from all methods (20). Regulators require product sponsors to document viral clearance in terms of a LRV (19).
In pH-based viral inactivation an acid (e.g., phosphoric acid) is added to a protein solution, which is held at the resulting low pH level for a validated time. A pH value of 3.9 or below is considered to be robust (20). This technique can inactivate enveloped viruses. The pH of the protein solution then is adjusted back to a physiological level, generally pH 7 or above. It is important to ensure that the low pH used to target viruses does not compromise the stability of a protein product itself. Low-pH viral inactivation generally is performed directly after protein A affinity capture chromatography, which uses low-pH buffers for column elution (21).
In solvent/detergent viral inactivation, a protein solution is incubated with a detergent and an organic solvent such as tri-(N-butyl) phosphate for a set time. After viral inactivation is complete, the solvent and detergents must be removed, typically by using a sorbent such as a polymer (19).
Viral Removal Methods
Size-exclusion techniques such as chromatography and viral filtration (nanofiltration) can remove nonenveloped viruses, which generally are chemically resistant and more challenging than enveloped viruses to remove (6). Even though column chromatography is not designed to eliminate viruses, it can remove both enveloped and nonenveloped viruses (22). However, the process is controlled by a number of operating parameters (e.g., temperature, flow rates, buffers, and wash volumes) that can affect the extent of viral reduction achieved. Thus, chromatography is a less favored approach than filtration (17).
Generally, nanofiltration is the method of choice for removing both small and large enveloped viruses as well as nonenveloped viruses. Membrane chromatography, however, is expanding in use because it uses virus-binding ligands along with ion-exchange adsorbers. In addition, it can operate at much higher flow rates than traditional column chromatography methods — which in turn helps speed up bioprocessing (19).
Regulatory agencies advise incorporating multiple orthogonal methods for viral clearance: independent methods with unrelated clearance methodologies (13). A successful viral clearance strategy can ensure that the chance of biopharmaceutical contamination by viruses is less than one in a million (17).
References
1 ICH 5A: Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. US Fed. Reg. 63(185) 1998: 51074; https://database.ich.org/sites/default/files/Q5A_R1_Guideline.pdf.
2 Merten OW. Virus Contaminations of Cell Cultures: A Biotechnological View. Cytotechnol. 39 (2) 2002: 91–116; doi:10.1023/A:1022969101804.
3 WHO Technical Report Series #978, Annex 3. Recommendations for the Evaluation of Animal Cell Cultures as Substrates for the Manufacture of Biological Medicinal Products and for the Characterization of Cell Banks. World Health Organization: Geneva, Switzerland, 2010; www.who.int/biologicals/vaccines/TRS_978_Annex_3.pdf?ua=1.
4 Klug B, Robertson JS, Condit RC. Adventitious Agents and Live Viral Vectored Vaccines: Considerations for Archiving Samples of Biological Materials for Retrospective Analysis. Vaccine 34 (51) 2016: 6617–6625; doi:10.1016/j.vaccine.2016.02.015.
5 Aranha H. Virus Safety of Biopharmaceuticals: Absence of Evidence Is Not Evidence of Absence. Contract Pharma 14 November 2011; www.contractpharma.com/issues/2011-11/view_features/virus-safety-of-biopharmaceuticals.
6 Challener CA. Viral Clearance Challenges in Bioprocessing. BioPharm Int. 27(11) 2014: www.biopharminternational.com/viral-clearance-challenges-bioprocessing.
7 Adair R. Control Viral Contaminants with Effective Testing. BioPharm Int. 30(10): 18–27; www.biopharminternational.com/control-viral-contaminants-effective-testing.
8 Xu Y, Brorson K. An Overview of Quantitative PCR Assays for Biologicals: Quality and Safety Evaluation. Dev. Biol. (Basel) 113, 2003: 89–98.
9 Braxton C. Next Generation Sequencing: The Utilization and Challenges for the Advancement of Viral Detection in the Biopharmaceutical Industry. Am. Pharmaceut. Rev. 15 August 2017; www.americanpharmaceuticalreview.com/Featured-Articles/341250-Next-Generation-Sequencing-The-Utilization-and-Challenges-for-the-Advancement-of-Viral-Detection-in-the-Biopharmaceutical-Industry.
10 An Introduction to Next-Generation Sequencing Technology. Illumina, Inc.: San Diego, CA, 2017; www. illumina.com/content/dam/illumina-marketing/documents/products/illumina_ sequencing_introduction.pdf.
11 Wetterstrand KA. The Cost of Sequencing a Human Genome. National Human Genome Research Institute, 10 July 2019; www.genome.gov/sequencingcosts.
12 5.2.3: Cell Substrates for the Production of Vaccines for Human Use. European Pharmacopoeia 9, 2017; www.edqm.eu/en/europeanpharmacopoeia-9th-edition.
13 CPMP/BWP/268/95. Note for Guidance on Virus Validation Studies: The Design, Contribution and Interpretation of Studies Validating the Inactivation and Removal of Viruses. European Medicines Agency: London, UK, 1996: www.ema.europa.eu/documents/scientific-guideline/note-guidance-virus-validation-studies-design-contribution-interpretation-studies-validating_en.pdf.
14 Stocking C, Kozak CA. Murine Endogenous Retroviruses. Cell Mol. Life Sci. 65(21) 2008: 3383–3398; doi:10.1007/s00018-008-8497-0.
15 De Wit C, Fautz C, Xu Y. Real-Time Quantitative PCR for Rerovirus-Like Particle Quantification in CHO Cell Culture. Biologicals 28(3) 2000: 137–148.
16 Farcet AMR, Kreil TR. Virus Susceptibility of Chinese Hamster Ovary (CHO) Cells and Detection of Viral Contaminations By Adventitious Agent Testing. Biotechnol. Bioeng. 106(4) 2010: 598–607; doi:10.1002/bit.2272.
17 Remington KM. Fundamental Strategies for Viral Clearance — Part 1: Exploring the Regulatory Implications. BioProcess Int. 13(1) 2015: 10–17. Part 2: Technical Approaches. BioProcess Int. 13(5) 2015: 10–16.
18 Noa E, et al. Downstream Processing: A Revalidation Study of Viral Clearance in the Purification of Monoclonal Antibody CB.Hep-1. BioPharm Int. 20 (1) 2007: www.biopharminternational.com/downstream-processing-revalidation-study-viral-clearance-purification-monoclonal-antibody-cbhep-1.
19 Sandle, T. Current Methods and Approaches for Viral Clearance. Am. Pharmaceut. Rev. September–October 2015: 1–4; www.americanpharmaceuticalreview.com/Featured-Articles/179320-Current-Methods-and-Approaches-for-Viral-Clearance.
20 Aranha H, Forbes S. Viral Clearance Strategies for Biopharmaceutical Safety, Part 2: A Multifaceted Approach to Process Validation. BioPharm 14(2) 2001: 43–54.
21 Shukla AA, et al. Evolving Trends in MAb Production Processes. Bioeng. Translat. Med. 2(1) 2017: 58–69; doi:10.1002/btm2.10061.
22 Zhang M, Miesegaes GR, Lee M. Quality By Design Approach for Viral Clearance By Protein A Chromatography. Biotechnol. Bioeng. 111(1) 2014: 95–103; doi:10.1002/bit.24999.
Further Reading
CPMP/BWP/269/95, Rev. 2. Note for Guidance on Plasma-Derived Medicinal Products. European Medicines Agency: London, UK, 2001; www.ema.europa.eu/en/documents/scientific-guideline/note-guidance-plasma-derived-medicinal-products_en.pdf.
Zhang J. Mammalian Cell Culture for Biopharmaceutical Production. Manual of Industrial Microbiology and Biotechnology, Third Edition. Baltz RH, Davies JE, Demain AL, Eds. American Society of Microbiology: Washington, DC, 2010: 157–178.
Shada Warreth is a senior bioprocessing trainer at the National Institute for Bioprocessing Research and Training (NIBRT), Foster Avenue, Mount Merrion, Blackrock, County Dublin, Ireland; 353-1-215-8136; [email protected].
You May Also Like






