Viral Clearance in a Downstream AAV Process: Case Study Using a Model Virus Panel and a Noninfectious SurrogateViral Clearance in a Downstream AAV Process: Case Study Using a Model Virus Panel and a Noninfectious Surrogate
Similarity in structures of adenoassociated virus serotype 8 (top) and minute virus of mice (bottom).
Photo credit: Protein Data Bank (https://www.rcsb.org)
Over the past decade, adenoassociated virus (AAV) vectors have become established as leading gene-delivery vehicles. In 2017, the pipeline for gene therapies included 351 drugs in clinical trials and 316 in preclinical development (1–4). As those candidates advance, significant efforts are being made in process development and manufacturing for viral vectors, with the overall goal of reducing process impurities while maintaining the highest possible process yield.
To address that goal, industry suppliers have developed innovative AAV-specific separation technologies. Thermo Fisher Scientific’s POROS CaptureSelect AAVX affinity resin provides a capture method for a number of natural and synthetic AAV serotypes irrespective of the expression system used to produce them. By leveraging a proprietary recombinant camelid antibody technology immobilized onto the highly permeable POROS backbone, the resin achieves a fine-tuned specificity for AAV recognition with an increased surface area and capacity for AAV binding. The significant impurity reduction benefits and rapid scalability of this affinity resin have led to its incorporation into several noted AAV downstream process designs.
BIA Separations (now part of Sartorius AG) has developed and commercialized CIMmultus QA monoliths, which have been cited in several AAV downstream processes for their ability to separate empty and full virus particles effectively. Monolithic supports represent a unique type of stationary phase for liquid chromatography, bioconversion, and solid-phase synthesis. Aside from increased processing speed, monolithic flow-through pores (channels) also provide easy access for large molecules, which supports both purification and depletion of nanoparticles such as plasmid DNA (pDNA) molecules and AAV particles.
One elusive aspect of AAV process development is viral clearance. As outlined in the ICH Q5A guidelines, viral clearance validation is a key regulatory requirement governing all recombinant biopharmaceuticals (5). According to these guidelines, the risks of viral contamination should be assuaged by a three-pronged approach: prevent, test, and remove. Over the past few decades, certain viral clearance strategies for monoclonal antibodies (MAbs), such as low-pH inactivation and nanofiltration, have become standard for most downstream processes. However, because AAVs are viruses themselves (within the family Parvoviridae), it may not be possible to apply the same purification strategies to them that have served so well in MAb processes. As a result, the gene-therapy industry may depend increasingly on chromatographic modes of separation to demonstrate sufficient viral clearance.
In the study reported herein, we addressed viral removal by performing scale-down–model spiking studies and measuring viral clearance using a clinically relevant AAV8 downstream purification process. The two-step chromatography process begins with affinity capture using POROS CaptureSelect AAVX affinity resin followed by anion-exchange polishing using a CIMmultus QA monolith.
We selected as spiking agents a wide range of viruses with different sizes, molecular makeups, and physiochemical properties. For DNA viruses, we used enveloped pseudorabies virus (PRV) and nonenveloped minute virus of mice (MVM). For RNA viruses, we used enveloped xenotropic murine leukemia virus-related virus (XMuLV) and nonenveloped reovirus type 3 (Reo-3). We also included human contagions hepatitis A (HAV) and herpes simplex virus 1 (HSV-1) based on a risk assessment of possible adventitious virus contaminations with a HEK293 human-derived producer cell line used for upstream production.
For the benefit of future process developers who may wish to perform similar viral clearance testing on their own purification process steps but lack the ability to conduct such resource-intensive studies, we assessed the MockV MVM kit from Cygnus Technologies, LLC, as an economical and rapid means to generate predictive MVM clearance data (6). We used the kit in parallel with MVM spiking experiments throughout our study.
Materials
All AAVs used for these viral clearance studies were produced at representative scale by a platform process at REGENXBIO Inc.
Production of Virus and Mock Virus Particles (MVPs): Viruses were propagated and purified by Texcell NA of Frederick, MD, according to standard protocols. Noninfectious MVM-MVPs were assembled after expression of the major MVM capsid protein (VP2) in a baculovirus expression-vector system (BEVS) using Spodoptera frugiperda 9 (Sf9) cells at Cygnus Technologies. Particles were purified with affinity and ion-exchange chromatography (IEC). Transmission electron microscopy (TEM) was used to verify MVP morphology, size, and concentration.
Chromatography Products: Thermo Fisher Scientific supplied prepacked, 5-mL POROS CaptureSelect AAVX affinity resin columns; BIA Separations provided scale-down CIMmultus QA monolith devices in 4-mL and 8-mL diameters. Control POROS resins were custom made by Thermo Fisher Scientific with identical base beads to the affinity resin used but incorporating either an alternative VHH ligand specificity (nonbinding for AAVs) or no VHH functionalized ligand.
Methods
Study Design: To assess the robustness of the affinity resin and monolith polishing step within REGENXBIO’s downstream process (7), we selected “center-point” and “worst-case” processing parameters for our viral clearance spiking experiments (Table 1). For each run, we spiked in-process AAV material with model viruses or MVM-MVPs to a target of 10.0 log10 MVM-MVP/mL and processed accordingly.
For affinity resin center-point and alternative-ligand runs, we loaded 150 mL of spiked material according to standard manufacturing load ratios and residence time. For worst-case conditions, we loaded 200 mL of spiked material (133% of the target) and decreased the flow rate to lengthen residence time to 170% of the center-point target. For the monolith polishing study, we applied center-point load volumes of 90 mL to 8-mL monoliths and 65 mL to 4-mL monoliths for worst case — except for the MVP- and XMuLV-spiked runs. For those, we loaded 45 mL and 65 mL onto 4-mL columns, respectively, for the center-point and worst-case conditions.
Samples were collected from each run during each step phase (flow-through, wash, and so on). We analyzed the virus samples immediately with a 50% tissue culture infectious dose (TCID50) assay or quantitative real-time polymerase chain reaction (qPCR). MVM-MVP samples were stored at –80 °C before Immuno-qPCR analysis (described below). From those results, we determined log reduction values (LRVs) using a standard calculation (5).
 During this study, we performed affinity-capture experiments to probe potential nonspecific binding interactions (Table 1). Both AAV-null (produced by pooling the flow-through fractions of previous AAVX runs) and AAV8-containing load materials were spiked with model virus, then affinity purified using center-point conditions and compared for viral clearance. Additionally, we evaluated interactions between viruses and base beads by performing AAV8 center-point runs using the POROS base matrix without a functionalized VHH ligand; we evaluated virus–VHH ligand interactions using a POROS resin with an alternative VHH ligand specificity to the Fc portion of MAbs that cannot bind AAVs.
During this study, we performed affinity-capture experiments to probe potential nonspecific binding interactions (Table 1). Both AAV-null (produced by pooling the flow-through fractions of previous AAVX runs) and AAV8-containing load materials were spiked with model virus, then affinity purified using center-point conditions and compared for viral clearance. Additionally, we evaluated interactions between viruses and base beads by performing AAV8 center-point runs using the POROS base matrix without a functionalized VHH ligand; we evaluated virus–VHH ligand interactions using a POROS resin with an alternative VHH ligand specificity to the Fc portion of MAbs that cannot bind AAVs.
We applied an orthogonal test method — surface plasmon resonance (SPR) — to confirm the specificity of the AAVX ligand for AAV. For that, a biotinylated AAVX VHH ligand was immobilized onto a detection surface so that binding sensograms could be generated by injection of free MVM-MVP or AAV.
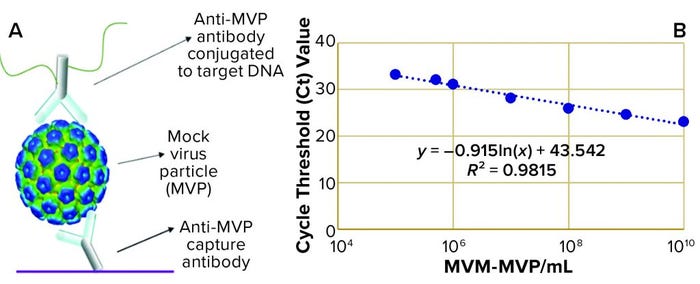
Figure 1: (a) Immuno-quantitative real-time polymerase chain reaction (Immuno-qPCR) assay; (b) immuno–qPCR standard curve
Analytical Assays and LRV Determinations: Texcell scientists quantified infectious titer of XMuLV using a validated plaque-forming infectivity assay. They quantified HAV, Reo-3, HSV-1, and MVM using validated TCID50 infectivity assays. PRV was quantified with a validated qPCR assay. From those titer determinations, we calculated LRVs by a standard method (5).
To analyze the concentration of noninfectious MVM-MVP within each sample, Cygnus Technologies scientists performed an Immuno-qPCR assay (Figure 1a) as described elsewhere (8). In brief, samples were added to microwells coated with an anti–MVM-MVP capture MAb. After incubation and washing, a DNA-conjugated anti–MVM-MVP detector MAb was added. Following another incubation and washing step, a dissociation buffer was added to each well for five minutes. Then 5 µL of sample was transferred from each well to a qPCR plate containing TaqMan primers/probes (Thermo Fisher Scientific) directed against the conjugated DNA. To determine the quantity of particles in unknown samples, threshold cycle (Ct) values were interpolated into a standard curve generated by including a 10-fold dilution series of a known MVM-MVP standard (Figure 1b). From those concentration values, we could calculate MVM-MVP LRVs for each experiment.
Results and Discussion
Viral clearance strategies for AAV downstream processes are limited by difficulty in performing viral inactivation and filtration steps without also inactivating or removing the AAV product. Therefore, chromatographic modes of separation are paramount to achieving the desired levels of removal for viruses of concern.
To complicate matters further, MVM (a small, nonenveloped virus used internationally as a model spiking agent) is a member of the same parvovirus family as AAV. Morphology and physicochemical properties (size, surface charge, and surface hydrophobicity) are similar in the two virus species. Because of those physicochemical similarities, a step optimized to bind and elute AAV through affinity or ion interactions also might bind MVM. That would result in poor removal of such viral contaminants — or conversely, a step optimized to remove MVM could compromise AAV yield as the product is washed or eluted away along with MVM.
With that in mind, in our study we wanted to elucidate whether viruses (including MVM) could be resolved from AAV through the combination of a POROS CaptureSelect AAV column and a CIMmultus QA monolith, thereby providing effective viral clearance.
Affinity Capture: Figure 2 summarizes the results from all viral clearance spiking experiments using POROS CaptureSelect AAVX affinity resin. At manufacturing center-point process conditions, effective viral clearance of ≥4 LRV was demonstrated for XMuLV, MVM, HAV, and PRV. The AAVX resin also contributed to clearing ≥2.5 LRV for Reo-3 and HSV-1. During worst-case–conditions testing, similar levels of clearance were observed for all model viruses tested. Taken together, this demonstration of robust viral clearance using POROS CaptureSelect AAVX affinity chromatography is consistent with the highly specific nature of the affinity interaction between AAVX resin and AAV vectors. Its high degree of specificity and capacity is mediated by the camelid VHH antibody ligands functionalized to the custom-designed base beads, which in combination provide high-affinity binding to AAV vectors while minimizing nonspecific interactions.
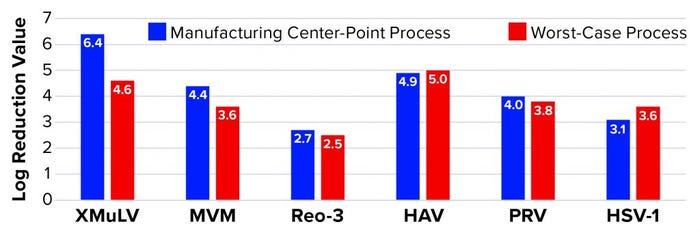
Figure 2: Viral clearance using affinity resin; see Table 1 for full virus names
Among the panel of six model viruses tested, MVM is potentially problematic to remove based on its similarity in size to AAV and high resistance to inactivation. Being a nonenveloped DNA virus that is a member of the same Parvoviridae family as AAV, MVM has a similar capsid structure and therefore potentially similar viral morphology and physicochemical properties (size, surface charge, surface hydrophobicity).
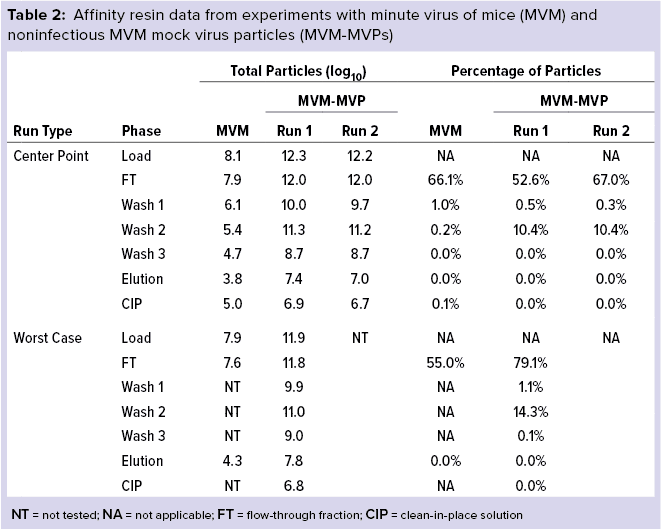
Table 2 details our results for MVM and MVM-MVP (the noninfectious MVM surrogate created by Cygnus Technologies as a biosafety-level 1 safe analytical tool). Most MVM initially loaded onto the column was contained within the flow-through fractions for both center-point and worst-case runs (66.1% and 55.0%, respectively). Similarly, most MVM-MVPs also were found within the flow-through fraction (52.6–67.0% and 79.1% for center-point and worst-case runs, respectively). Affinity wash steps did little to strip the column further of MVM (0.2% for center point); moderate quantities of MVM-MVPs were removed (10.4% and 14.3% for center-point and worst-case runs, respectively). Small quantities of both MVM and MVM-MVPs were found in the elution fractions collected.
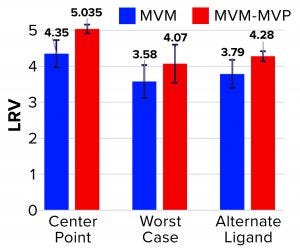
Figure 3: Log reduction value (LRV) determinations for affinity resin runs spiked with minute virus of mice (MVM) and noninfectious MVM mock virus particles (MVM-MVPs)
From MVM center-point and worst-case runs, 3.8 log10 and 4.3 log10 total particles were detected in the elution, respectively, leading to LRV calculations of 4.35 ± 0.38 and 3.58 ± 0.46 (Figure 3). That difference in LRV probably can be attributed to the influence of process parameters (load ratio and residence time) on virus–ligand interactions. As predicted, a higher load ratio and residence time yielded nearly a 1.0 log10 decrease in MVM clearance. For MVM-MVP center-point and worst-case runs, 7.0–7.4 and 7.8 log10 total particles were determined, respectively, giving LRV calculations of 4.91–5.16 and 4.07. Thus, similar clearance results were achieved for both particle types at each condition, and the trend in reduced clearance seen for MVM could be monitored through the use of MVM-MVP. Figure 3 also shows LRV results for a center-point run using an alternative base matrix.
Overall, those results demonstrate the high selectivity of the POROS CaptureSelect AAVX affinity resin, which can differentiate between the surface epitopes of AAV and the evolutionarily similar virus MVM. Such high specificity enables the resin to partition those two particle types from a heterogenous mixture containing both of them. Our data also reveal comparable results between MVM and MVM-MVP, demonstrating the utility of MVM-MVP as a spiking/analysis tool for process development and characterization.
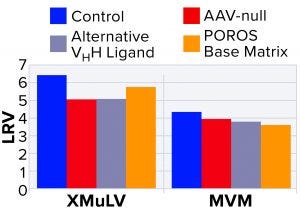
Figure 4: Characterization of nonspecific interactions based on log reduction values (LRVs) for xenomorphic murine leukemia virus (XMuLV) and minute virus of mice (MVM)
To probe nonspecific binding, a more detailed interaction study was performed using MVM and XMuLV, which are the two most commonly used model viruses for viral clearance spiking studies (9). To probe virus–AAV interactions, we performed an AAV-null run wherein the spiked virus load was devoid of AAV8 product. As Figure 4 shows, the null run demonstrated similar performance to the manufacturing control run for both MVM and XMuLV, indicating that the presence of AAV8 had minimal effect on clearance of model viruses.
Next, to probe interactions among viruses and VHH ligands or POROS base beads, we used two control resins designed by Thermo Fisher Scientific. The first control was an AAVX-like resin with an identical base bead but a functionalized VHH ligand with an alternate specificity that cannot bind AAV. The second control was POROS CaptureSelect AAVX resin without a functionalized VHH ligand. Using these control resins, we observed similar viral clearance levels to those of the center-point run using unmodified POROS CaptureSelect AAVX affinity resin, which indicates that minimal interactions occurred between viruses and the ligand or base beads. These results strongly indicate that model viruses show no nonspecific binding to either the VHH ligand or to the POROS base bead — and that interactions between those viruses and the AAV product are minimal.
Figure 5: Binding selectivity of camelid VHH antibody-fragment affinity ligand analyzed by surface plasmon resonance (SPR)
SPR results demonstrated that the AAVX VHH ligand bound to the injected AAV1, but not to MVM-MVP (Figure 5). For an experimental control, the binding signal was recovered when AAV was spiked back into a 0.1-µg/mL MVM-MVP background. These data suggest that the presence of virus particles (infectious or otherwise) does not interfere with the ability of AAV to bind to the specific AAVX VHH ligand.
Monolith Polishing: Tables 3–9 show complete viral clearance results (including MVM-MVP) from our CIMmultus QA spiking experiments. During each experiment, we captured flow-through, three pre-AAV elution peaks, two postproduct peaks, and a strip fraction along with the load and main AAV elution pool. As the data show, no virus (or MVM-MVP) was detected in any fraction other than the strip for either center-point or worst-case conditions. That indicates complete clearance for all virus types in this downstream AAV process step. Viruses also were undetectable in the flow-through, pre– and post–main-peak collections.
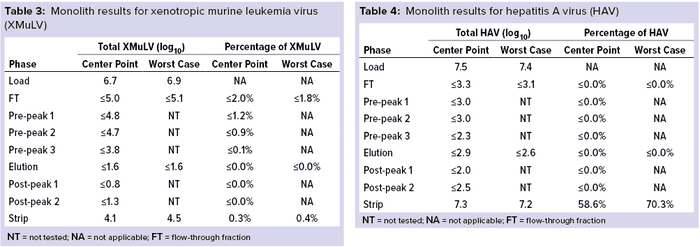
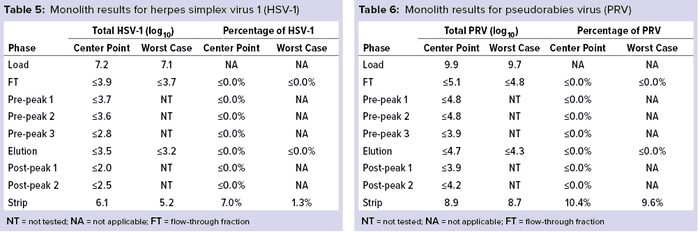
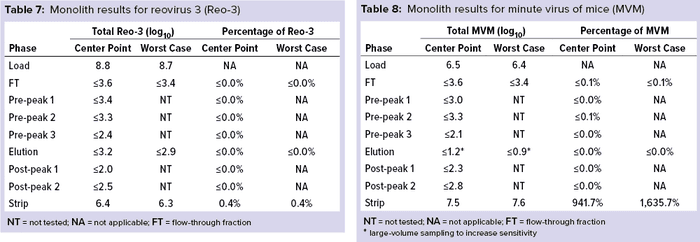
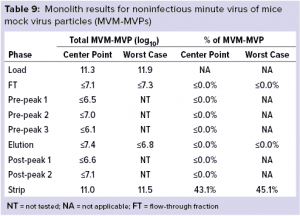 Virus titers within the strip fractions were significant but differed by virus. The amount of MVM detected in the strip fraction was greater than the overall challenge, which could indicate interference with the assay. In some cases, the mass-balance of total virus detected within the collected fractions did not equate to the amount of virus challenged. That may be attributable to (partial) degradation of the virus by the stripping agent and/or to using the stripping agent for too short a time to elute all the virus.
Virus titers within the strip fractions were significant but differed by virus. The amount of MVM detected in the strip fraction was greater than the overall challenge, which could indicate interference with the assay. In some cases, the mass-balance of total virus detected within the collected fractions did not equate to the amount of virus challenged. That may be attributable to (partial) degradation of the virus by the stripping agent and/or to using the stripping agent for too short a time to elute all the virus.
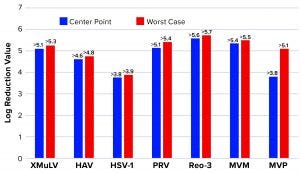
Figure 6: Monolith log reduction values (LRVs); see Table 1 for full virus names
Figure 6 shows LRVs from each experiment. The monolith offered effective removal for a wide range of physicochemically distinct viruses. In addition, the LRV data demonstrate comparability between MVM and MVM-MVP clearance at both center-point and worst-case conditions.
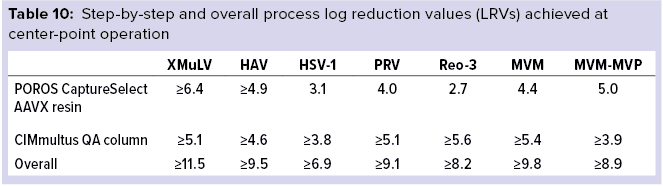
Final Results: Table 10 lists overall process LRVs achieved after using both POROS CaptureSelect AAVX affinity resin and CIMmultus QA monolith polishing steps operated at center-point manufacturing conditions.
An Accurate and Economic Prediction Model
We sought to determine whether effective viral clearance could be achieved through chromatographic methods in an AAV purification process. Through spiking studies using a broad and inclusive panel of viruses, we determined that chromatographic modes of separation indeed can provide an effective viral clearance strategy. Both POROS CaptureSelect AAVX affinity resin and CIMmultus QA anion-exchange monoliths demonstrated superb ability to reduce viral levels and contribute to high overall process LRVs.
During this study, we also sought to determine whether a biosafety-level 1 compliant, noninfectious mock MVM particle could serve as an accurate surrogate for predicting MVM removal. High correlation between the MVM and MVM-MVP results obtained throughout this study suggest that such an approach could provide an accurate and economic model for predicting the viral clearance efficacy of other AAV chromatographic separation techniques.
References
1 Rininger J, Fennell A, Schoukroun-Barnes LR. AAV Vector Manufacturing Platform Selection and Product Development. BioProcess Int. December 2019 sponsored insert; https://bioprocessintl.com/sponsored-content/aav-vector-manufacturing-platform-selection-and-product-development.
2 Scull L. Gene Therapy Pipeline Overview. ATHN Data Summit: Chicago, IL, 25–26 October 2018; https://alliancerm.org/wp-content/uploads/2018/10/Gene-Therapy-Pipeline-Overview_FINAL-2.pdf.
3 Ginn SL, et al. Gene Therapy Clinical Trials Worldwide to 2017: An Update. J. Gene Med. 20(5) 2018: e3015; https://doi.org/10.1002/jgm.3015.
4 Lloyd I, et al. Pharma R&D Annual Review. Pharmaprojects: London, UK, February 2017; https://pharmaintelligence.informa.com/~/media/Informa-Shop-Window/Pharma/Files/PDFs/whitepapers/RD-Review-2017.pdf.
5 ICH Q5A: Viral Safety Evaluation of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. US Fed. Reg. 63(185) 1998: 51074; http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/
Guidances/ucm073454.pdf
6 Orchard JD, et al. Using a Noninfectious MVM Surrogate for Assessing Viral Clearance During Downstream Process Development. Biotechnol. Prog. 36(1) 2020: e292; https://doi.org/10.1002/btpr.2921.
7 Goldfarb M, et. al. Downstream Purification of Adeno-Associated Virus for Large-Scale Manufacturing of Gene Therapies. Cell Gene Ther. Ins. 6(7) 2020: 955–963.
8 Cetlin D, et al. Use of a Noninfectious Surrogate to Predict Minute Virus of Mice Removal During Nanofiltration. Biotechnol. Prog. 34(5) 2018: 1213–1220; https://doi.org/10.1002/btpr.2694.
9 Herbig K, et al. Modeling Virus Clearance: Use of a Noninfectious Surrogate of Mouse Minute Virus As a Tool for Evaluating an Anion-Exchange Chromatography Method. BioProcess Int. 17(5) 2019: 34–40; https://bioprocessintl.com/downstream-processing/viral-clearance/modeling-virus-clearance-use-of-a-noninfectious-surrogate-of-mouse-minute-virus-as-a-tool-for-evaluating-an-anion-exchange-chromatography-method.
Corresponding author Michael Winkler ([email protected]) is director of downstream process development and validation, Mikhail Goldfarb* was a principal scientist in process development, Shaojie Weng is a scientist II, Jeff Smith is a senior associate scientist, and Susan Wexelblat is a CMC technical writer at Regenxbio Inc., 9600 Blackwell Road, Suite 210, Rockville, MD 20850. John Li is a staff scientist, Alejandro Becerra is a principal applications scientist, and Sandra Bezemer and Kevin Sleijpen are scientists at Thermo Fisher Scientific, 35 Wiggins Avenue, Bedford, MA 01730. Aleš Štrancar is chief executive officer, Sara Primec is sales and product manager, and Romina Zabar is head of quality assurance at BIA Separations, Mirce 21, SI-5270 Ajdovščina, Slovenia. April Schubert is director of business development for North America, and Akunna Iheanacho is scientific director at Texcell, 4991 New Design Road, Frederick, MD 21703. David Cetlin is senior director of R&D at Cygnus Technologies LLC, 4332 Southport-Supply Road SE, Southport, NC 28461.
*Goldfarb is now director of protein purification at Arcellx, Inc.
You May Also Like






