Polishing a Monoclonal Antibody (MAb) Using Capto™ adhere ImpRes in Bind and Elute ModePolishing a Monoclonal Antibody (MAb) Using Capto™ adhere ImpRes in Bind and Elute Mode

A platform approach for MAb purification is desirable as it saves both time and money in process development. GE Healthcare Life Sciences’ MAb toolbox employs protein A chromatography media (resins) as well as a wide range of options for polishing purification steps. One of these options is Capto adhere ImpRes, a cost-effective and flexible multimodal anion exchange chromatography medium designed for high-resolution polishing of MAbs. The medium allows operation in either bind-elute (B/E) or flow-through (FT) modes.
Here we describe a workflow for method development of a polishing step for a MAb in B/E mode using Capto adhere ImpRes.
Screening for Optimal Binding Conditions
The influence of pH and NaCl concentration on static binding capacity and dynamic binding capacity of Capto adhere ImpRes was determined in PreDictor™ 96-well filter plates and by design of experiment (DoE) in Tricorn™ 5/50 columns, respectively. The results show that the highest binding capacities were obtained at high pH and low salt concentration, while a shorter residence time reduced the capacity (Figure 1). Binding of the MAb, purified from CHO cell supernatant by protein A, to Capto adhere ImpRes was therefore performed in 40 mM sodium phosphate, pH 7.8.
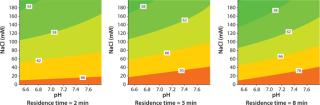
Figure 1: ()
Screening for Optimal Elution Conditions
Determination of yield at different elution conditions was performed in PreDictor plates. The highest yield was obtained at low pH and low salt concentration (Figure 2).
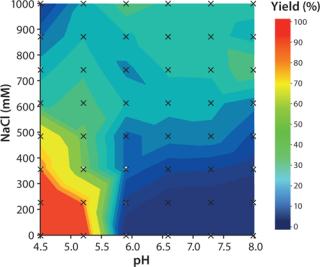
Figure 2: Column: Tricorn 5/50, column volume ~1 mL
Medium: Capto adhere ImpRes
Sample: MAb partially purified by protein A chromatography
Sample Load: 43.4 mg MAb/mL chromatography medium
Start Buffer: 40 mM sodium phosphate, pH 7.8
Elution Buffer: 20 mM sodium phosphate, 20 mM citrate, pH 4.0 (blue curve); 20 mM sodium phosphate, 20 mM citrate, 100 mM NaCl, pH 4.0 (green curve)
Gradient: 0–100% elution buffer in 20 CV
Residence Time: 4 minutes
System: ()
Gradient elution by pH was evaluated with or without addition of NaCl. Addition of NaCl in the elution buffer resulted in slightly lower elution pH, lower aggregate content, and a broader elution peak than elution buffer without NaCl (Figure 3).
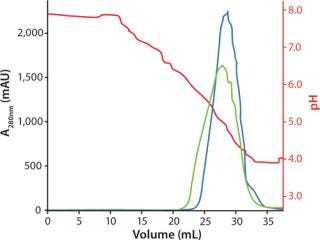
Figure 3: ()
Conditions for step elution were further investigated using DoE, varying sample load between ~50% and 70% of the dynamic binding capacity. Verification of the design model was finally performed in a Tricorn 5/50 column.
Results and Conclusion
The responses from the design model show that the only significant factor was elution pH. Higher elution pH resulted in lower yield and lower aggregate concentration. The model suggested an elution pH of 4.5 (0 M NaCl) and a sample load of 70% of DBC (╛50 mg/mL).
Column verification of the design model shows that the obtained result was in good agreement with the expected. The aggregate content was reduced from 2.5% down to 0.8% with high yield. A more detailed description of this case study is given in the Application note 29-0273-38 (1).
About the Author
Author Details
Anders Ljunglof is a scientist, and Kristina Nilsson-Valimaa is a senior research engineer at GE Healthcare Bio-Sciences AB, Björkgatan 30, 751 84 Uppsala, Sweden.
REFERENCES
1.) Application Note 29-0273-38, and AA Edition. 2013.Polishing of Monoclonal Antibodies Using Capto adhere ImpRes in Bind and Elute Mode, GE Healthcare, Upssala.
You May Also Like






