Validation of Adventitious Virus Removal By Virus FiltrationValidation of Adventitious Virus Removal By Virus Filtration
Regulatory bodies around the world expect downstream purification processes to demonstrate robust clearance of model adventitious viruses in time for execution of phase 3 clinical trials and product licensure (1,2,3). Model viruses selected for these studies should represent a diversity of viral physicochemical properties, and the clearance methods applied should include orthogonal mechanisms such as clearance based on size alongside chemical inactivation. Virus filtration is a critical unit operation used in numerous purification processes of monoclonal antibodies (MAbs), recombinant proteins, and plasma-derived biopharmaceuticals.
PRODUCT FOCUS: ANTIBODIES AND OTHER RECOMBINANT PROTEINS
PROCESS FOCUS: DOWNSTREAM PROCESSING
WHO SHOULD READ: MANUFACTURING, PROCESS DEVELOPMENT, AND ANALYTICAL PERSONNEL
KEYWORDS: VIRAL SAFETY, MMV/MVM, XMULV, FILTRATION, SCALE-UP
LEVEL: INTERMEDIATE
Virus filtration unit operations have been shown to be scalable, robust, and reproducible (4, 5). Initial sizing of membranes usually involves a Vmax study on a small-scale filter followed by linear scale-up maintaining a constant ratio of process volume per square meter area. However, the ability of a virus filter to clear viruses is ultimately determined by validation studies involving virus-spike and clearance experiments. A representative feedstock is spiked with a preparation of model virus, this feedstock is passed through the filter, and viral clearance is determined after measuring recovered levels in filtrate using qualified/validated scale-down systems.
Because the level of impurities in a viral spike is much higher than in a nonspiked feed, significant degradation in volumetric capacity of a virus filter is sometimes observed when processing a spiked load (4). It has been noted that the ability of virus filters to clear small parvoviruses may be degraded when their membranes are fouled, as shown by accelerated decays in flow when using spiked loads (6, 7). This raises a question of whether virus filter sizing for routine processing should be based on capacity as determined using nonspiked feed or the more limited capacity obtained with a spiked feed during process validation (which, in most cases, is significantly smaller).

Figure 1:
Here we present a novel procedure for approaching the validation of adventitious virus removal while successfully resolving those issues. It must be pointed out that this method is tested only in certain virus filters from Millipore Corporation (www.millipore.com) — but these particular filters are widely used in the industry, so the method should have broad application.
Materials and Methods
Optiscale-25 devices with Viresolve NFP membranes (3.5 cm2) were purchased from Millipore. Such filters are designed to be used in normal flow mode (dead-end mode) and can be operated at constant pressure or constant flow. Optiscale-40 devices with Viresolve prefilters (5.0 cm2) were also purchased. They combine charged-modified depth-filtration media with a 0.1-µm nominally rated cellulose membrane layer at the bottom.
Load Material and Virus Assays for the Study: Load material used for this study came from clinical manufacturing runs using an intermediate product pool, with a concentration of 4–8 mg/mL at pH 7.5 and conductivity of 8–10 mS/cm.
Viruses used for this study are listed in Table 1 along with their physical properties. We assayed our load samples for cytotoxicity and interference of xenotropic murine leukemia virus (XMuLV) and murine minute virus (MMV) infectivity in PG-4 (feline S+L−) cells and 324 K cells, respectively. Cytotoxicity, interference, and infectivity assays were performed by BioReliance Corporation (www.bioreliance.com). For validation spiking studies, virus titers were 1.0 × 107.7 particles/mL for XMuLV and 1.0 × 107.9 particles/mL for MMV. For the development study, virus titer was 1.6 × 107 particles/mL. A viral spike volume of 0.05% (v/v) relative to the entire filtrate volume (v/v) was used in all spiking studies. LRV data at a specific percent of flow delay came from instantaneous samples (2 mL) collected from the permeate. We determined flow decay using this equation:
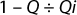
where Qi = initial flow rate at the start of the filter challenge, and Q = flow rate at the time the sample was taken.
Table 1: Viruses used in this study
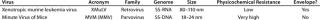
Table 1: Vi ruses used in this study ()
Experimental Method
Vmax Test Procedure: We used Optiscale devices with an effective membrane area of 3.5 cm2 and carried out our trials under a constant feed pressure of 30 psi. After a filter was connected to the set-up, buffer was added to the feed vessel, which was then pressurized to the test pressure. Then the buffer was allowed to flow through the membrane. We calculated buffer permeability values to ensure that the entire filter area was used. Next, the buffer was drained from the pressure vessel, and our protein solution was then added to the pressure vessel. The feed vessel was pressurized to the test pressure, and we recorded the weight of filtrate collected over time.
For some experiments, a Viresolve prefilter (VPF) was used in line with the Viresolve NFP filters in a similar ratio of membrane areas as used in offline experiments. For offline experiments the feed was prefiltered first through VPF, then loaded onto the NFP filter. For the latter, we maintained residence time through the VPF the same as for the downstream NFP filtration.
Results and Discussion
Effect of Load Conditions on NFP Capacity and Benefit of VPF: Preliminary studies have shown that changes in load protein concentration, hold time, and storage conditions (e.g., fresh load or previously frozen load) significantly affected the capacity of NFP filters. Their capacity was reduced at high protein concentrations, with long load hold times, and when using load that was previously frozen (data not shown).
We tested the VPF for its effect on capacity of the N
FP filters. In previous studies it was shown to improve their performance (8). In those experiments, prefilters enhanced the capacity of NFP filters and made their performance more reproducible under various load conditions including a freeze–thaw cycle or an extended load hold time (data not shown). In addition to reducing costs by providing greater capacity, such results enable implementation of a possible hold step for virus filter loads and also facilitate process validation studies because they are typically done on materials that have been previously frozen.
Effect of Virus Spike on Filter Capacity: Current practice for sizing virus filters is based on volumetric throughput achieved during virus spiking studies. Although viral clearance experiments are designed to reproduce actual process conditions at small scale, it is clear that introduction of virus spike into a representative protein solution dramatically decreases filter capacity because of fouling.
Figure 1 shows that capacity of our filter was greatly reduced as measured by Vmax testing when XMuLV was introduced into a MAb solution. A 0.05% (v/v) spiked load of XMuLV gave relatively higher capacity than did 0.2% and 1% spike loads, but it was decreased more than fivefold when compared with nonspiked feedstock. Data shown suggest that the quality of virus stocks used in spiking studies can significantly affect virus filter performance, potentially making scale-down studies less representative of a large-scale manufacturing process, which may lead to significant oversizing of virus filters. An effect of virus stock quality on filter validation has been reported (9). So we investigated alternative approaches to address this issue.
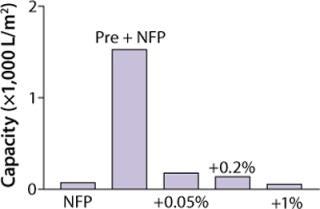
Figure 1: ()
Comparing Approaches: Preliminary experiments were performed in development to evaluate the affect of virus spike on Viresolve NFP filter capacity when using a “run-and-spike” approach. In that approach, nonspiked material is processed through a filter to beyond its target capacity. The filter is then challenged with a small volume of the same feedstock that has been spiked with virus. Table 2 shows results from a development run demonstrating that at a capacity beyond the normal target range (300–500 L/m2) the virus filter can clear more than three logs of a spiked model retrovirus.
Table 2: Run-and-spike approach using XMuLV in an R&D run

Table 2: Ru n-and-spike approach using XMuLV in an R&D run ()
Table 3 shows data comparing validation experiments using that conventional method with the “run-and-spike” approach. For the former experiment, we spiked 0.05% (v/v) XMuLV to the initial load, whereas for the latter, we spiked 0.05% XMuLV relative to the entire filtered volume into the remaining load so that the concentration of virus spike in this smaller volume was 0.5% (v/v). In both experiments, exactly the same amount of virus was processed over the filter — but in different spiked volumes. The results show a capacity of 567 L/m2 for the tested MAb pool using the run-and-spike method, whereas implementing the conventional method gave 276 L/m2, an over twofold increase of capacity.
Table 3: NFP capacity and XMuLV clearance using spike-and-run and run-and-spike approaches from validation runs

Table 3: NF P capacity and XMuLV clearance using spike-and-run and run-and-spike approaches from validation runs ()
We observed the level of virus reduction for both approaches to be comparable. In our validation study, the spike-and-run experiment also served as a control to ensure that fouling the VF membranes in the case of a more concentrated virus load in run-and-spike is not a concern because LRV data were comparable from both scenarios. The absolute LRV value was different in development and validation runs because of multiple factors (e.g., assay volume and viral titer used to spike).
Flow Decay and LRV: It has been reported that viral clearance capacity with small viruses (e.g., MMV) decreases as flow decays using the conventional spike-and-run method (3, 7). So it becomes critical to understand the specific nature of the effect of flow decay on a specific filtration step. It has been reported that LRV depends only on the degree of flow decay or plugging regardless of throughput or protein concentration (7).
Our investigation of the effect of flow decay showed that this phenomenon was observed using both the run-and-spike method and the spike-and-run approach (Table 4). Conditions that led to more than 90% decay from the initial flow rate still resulted in over four logs of MMV clearance. The results in Table 4 show similar patterns of flow decay and LRV for both types of experiments from an actual validation run. The advantage of using a run-and-spike approach is that total throughput (and therefore the capacity that can be validated) increases about twofold. Comparability of the LRVs in both scenarios suggests that fouling of VF membranes with a more-concentrated virus load in run-and-spike testing is not a concern.
Table 4: NFP capacity and MMV clearance using spike-and-run and run-and-spike approaches from validation runs

Table 4: NF P capacity and MMV clearance using spike-and-run and run-and-spike approaches from validation runs ()
Test Conclusions
Conventional approaches to validation of virus filtration steps may lead to conflicting capacity data when using nonspiked loads and virus-spiked loads in a validation study. Typically, the capacity observed in spiked samples is considerably lower because of fouling. If the capacity that can be validated is used to determine the appropriate amount of filter needed, it is likely that the recommended amount will be significantly more than necessary when filters are challenged routinely with nonspiked loads. We developed a run-and-spike approach that demonstrated efficient viral clearance while preserving the large process capacity inherent in these steps.
Along with comparable data from a spike-and-run experiment, run-and-spike LRV data allows for validation of a viral clearance claim. Use of the run-and-spike method also allows a claim of volumetric capacity significantly greater than that achieved using a classical spike-and-run approach.
During virus filtration, filter fouling usually results in flux loss. It is now understood that, as this membrane fouling occurs, the effectiveness of removing small viruses from feedstock may also degrade. In our studies, we observed this phenomenon using both the run-and-spike and spike-and-run approaches. In both cases, it is critical to understand the dynamic at work to make best use of the virus filtration step. It may be advantageous to routinely monitor process flow decay during virus filtration to ensure that claimed levels of small virus removal are achieved.
We found that a combination of validating the impact of flow decay on LRV at small scale (validation to a percentage of flow decay) with a run-and-spike testing lowered costs by reducing the amount of required virus filter area. In implementing this approach at commercial scale, the decrease in flux during execution of this step is continuously monitored to ensure that the step is operating under conditions that afford the potential for maximum virus clearance. The procedures outlined here have been reviewed, discussed, and accepted as suitable by regulatory bodies as part of a virus validation package to be included in an application for product licensure.
REFERENCES
1.) 1998. ICH Harmonized Tripartate Guideline: Q5A Viral Safety of Biotechnology Products Derived from Cell Lines of Human or Animal Origin. Fed. Reg. www.ich.org/LOB/media/MEDIA425.pdf 63:51074.
2.) CBER 1997. Points to Consider in the Manufacture and Testing of Monoclonal Antibody Products for Human Use, US Food and Drug Administration, Rockville.
3.) Kern, G, and M. Krishnan. 2006. Virus Removal By Filtration: Points to Consider. BioPharm Int. 19:32-41.
4.) PDA Virus Filter Task Force 2005. PDA Technical Report No. 41: Virus Filtration. PDA Journal 59.
5.) Ireland, T. 2004. Viral Filtration of Plasma Derived Human IgG: A Case Study Using Viresolve NFP. BioPharm Int..
6.) Atkinson, EM. 2003.. Effects of Volumetric Throughput and Membrane Flow Decay on Virus Retention: Ensuring Higher Safety Levels for Virus Reduction Filtration Processes.
7.) Genest, P. 2006. An Improved Method for Virus Filter Qualification and Implementation. BioProcess Int. 4:44-47.
8.) Ireland, T, G Bolton, and M. Noguchi. 2005. Optimizing Virus Filter Performance with Prefiltration. BioProcess Int. 3:S44-S47.
9.) Cabatingan, M. 2005. Impact of Virus Stock Quality on Virus Filter Validation. BioProcess Int. 3:S39-S43.
You May Also Like






